Applications:
 |
 |
 |
Direct Access
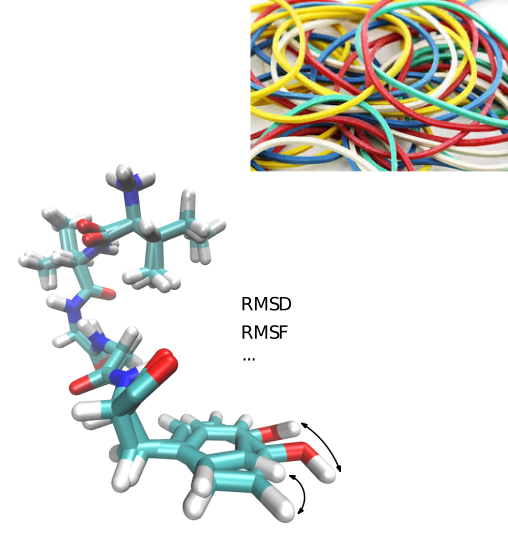
Space Filling Models
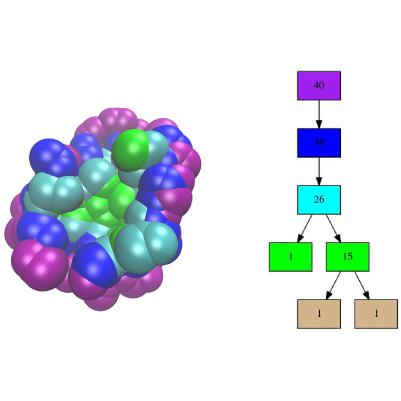
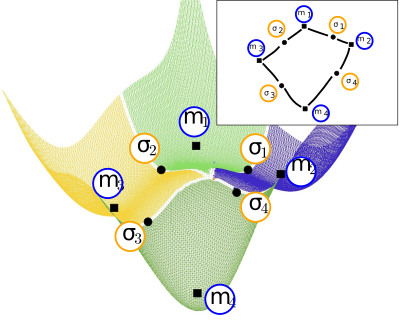
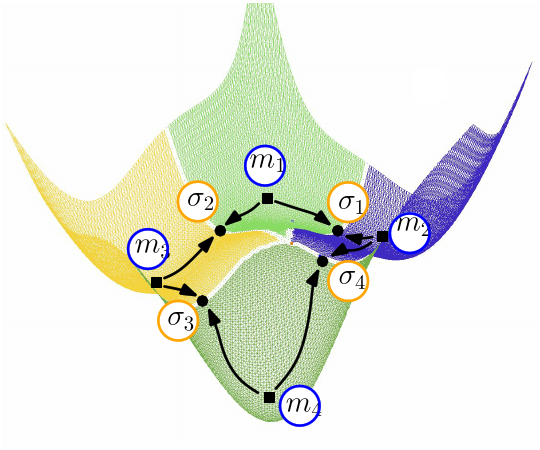
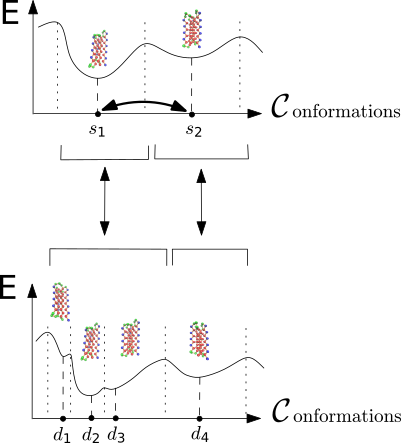
Conformational Analysis
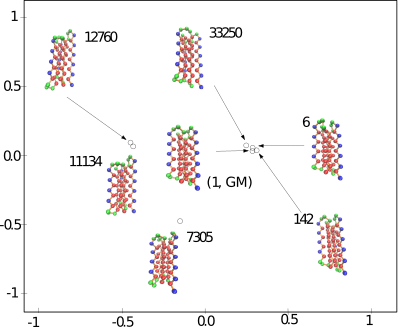
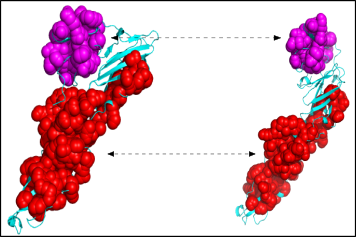
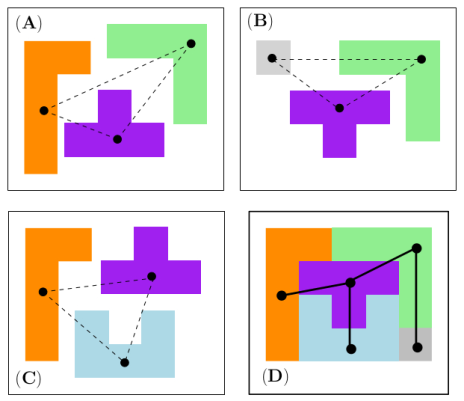
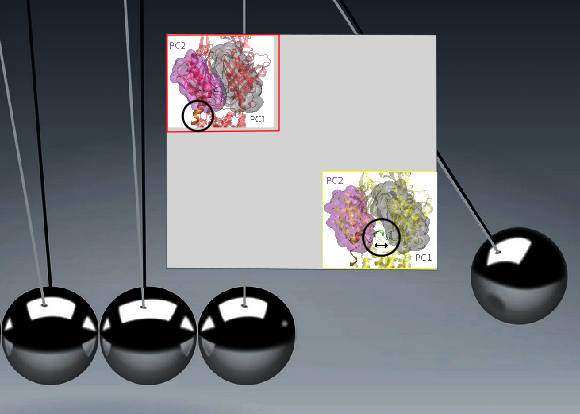
Large Assemblies
|
|
|
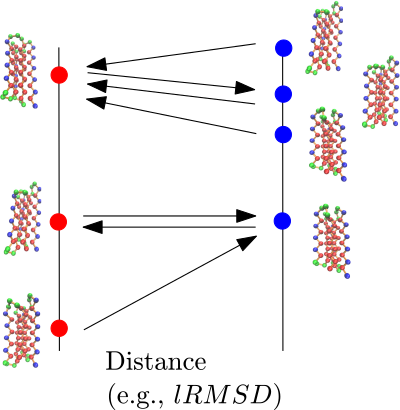
Integrated Analysis
|

|
|
|
|
|
|
|
|
|
|
|
|
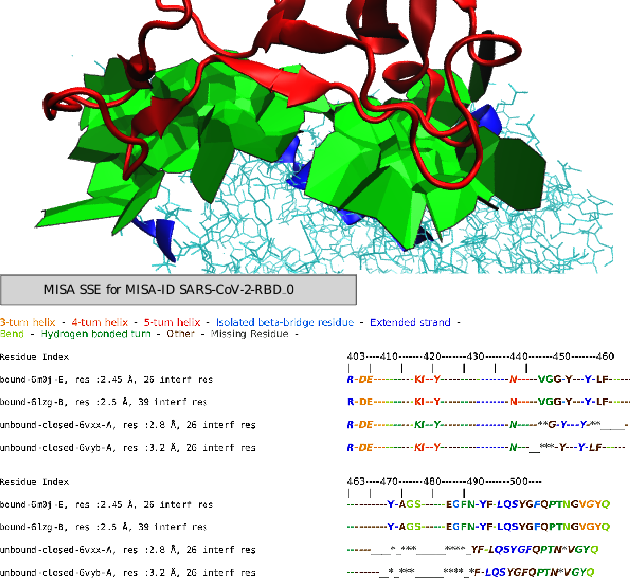
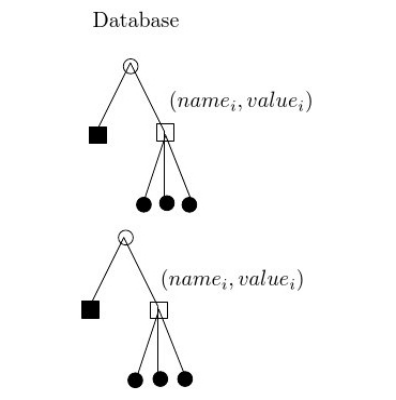
Data Management
|

|
|
|
|
|
|
|
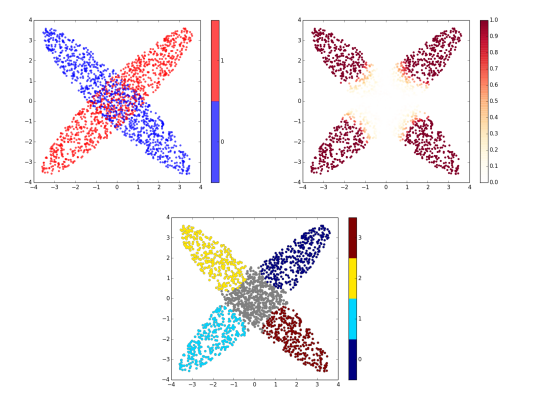
Data Analysis
|
|
|
|
|