 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|

Authors: R. Tetley and F. Cazals
Goals. In this package, we consider the problem of protein function annotation and prediction from structural and sequence information. In short: given a set of polypeptide chains involved in the same function, we wish to:
These questions are especially challenging when the sequences associated with the structures provided have low sequence identity – say < 20%. Our solution to this problem uses the identification of structurally conserved motifs, see [45] and Structural_motifs.
In a nutshell, given two proteins, a structural motif consists in two sets of a.a., one on each structure, such that the 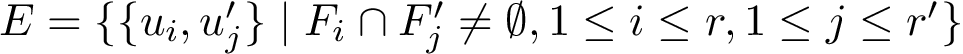
Overview of the method. Given a (small) set of structures, our method consists of three main steps:
Note that combining sub-sequences and whole sequences is meant to bias the MSA towards the important regions, while yet retaining information on linkers connecting these regions.
In the following, we detail the three steps just sketched.
Given two proteins, a structural motif consists in two sets of a.a., one on each structure, such that the 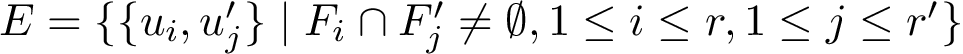
Note that two proteins typically generate several motifs, which can be filtered using motif inclusion and statistical significance. See the package Structural_motifs for the details.
Suppose the previous step yields a set 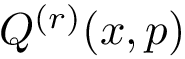
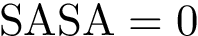
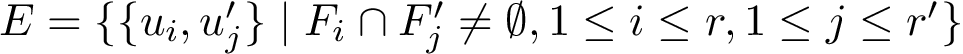
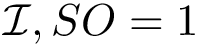

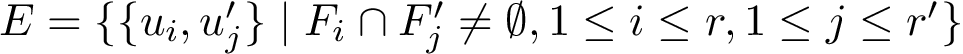
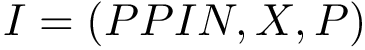
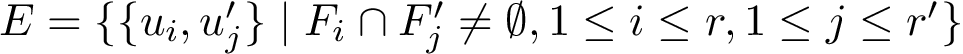
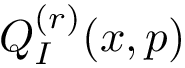
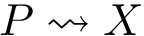
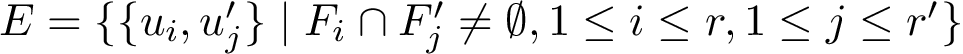
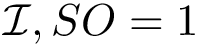
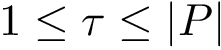
 |
Motifs and PCS for the Rift Valley Fever virus class II fusion protein a) The structure of the Rift Valley Fever virus class II fusion protein. Residues that belong to a motif which have a b) The sequence associated to the structure on the left. Residues that belong to a motif which have a c) The previously defined |
From the previously defined 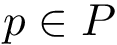
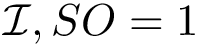
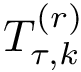
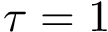
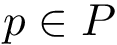
The multiple sequence alignment is performed using the Clustal Omega algorithm ([176]), or alternatively, the Muscle algorithm ([89]).
From the hybrid multiple sequence alignment, we build hidden Markov models. These models are used to query databases (notably the 
This step is performed by using the HMMER_Wrapper package.
Querying 
For a given hit, we use the DB_manipulator package and its DB_hits_manager.py module to recover the taxonomic information from the NCBI Taxonomy database and its fasta sequence. Each hit can be filtered by an e-value which assesses its significance.
To filter results, we annotate the sequence through the Protein_sequence_annotator package.
We are now able to filter the results through a selection of taxonomic or sequence criterions (see Protein_sequence_annotator and DB_manipulator for more details).
After performing Step 3, and filtering the hits, we obtain a set of protein sequences. This set can be used to bootstrap the process: we add these new sequences to the initial set and build a new HMM. We use this new HMM to perform a new query.
We provide two executables:
The main input of sbl-FunChaT-step-one.py is a folder containing the pdb files for each of the scrutinized structures.
Note that the executable makes use of the Batch_manager package. Default options are instantiated for Structural_motifs in the FunChaT-default.spec file. The user should take the time to fine tune these options.
A computation is launched as follows:
> sbl-FunChaT-step-one.py -p data/pdb -s data/FunChaT-default.spec -o results --parallel 4
| File Name | Description |
| PDB File | A .pdb file containing one of the processed structures. The script should be directed to a folder containing all PDB files of analyzed structures. |
| Spec File | A .spec file containing the specification of the run. |
The output consists of a list of folders each containing the output files of the Structural_motifs package.
The main input of sbl-FunChaT-step-two-and-three.py is a folder containing the results of Structural_motifs. The results of each pairwise comparison are bundled in a .xml file. The file for each of these comparisons should be contained in an independent folder with the standard name sbl-structural-motifs-*-pdb-fileStructure1-pdb-fileStructure2 (as per the Batch_manager default nomenclature).
Additionally, the user should provide a file containing, for each structure: its full fasta sequence, its name, a residue sequence number range corresponding to the residues contained in the sequence. (See example provided in the second line of the table below.)
A computation involving steps 2 and 3 is launched as follows:
> sbl-FunChaT-step-two-and-three.py -r results -f data/sequences.fasta -d ALL -t 0.8 -o results/hmm_results -a $UNIPROT_FASTA_DIR -u $UNIPROT_DIR -n $NCBI_DIR
Or if a user would prefer to only perform step two:
> sbl-FunChaT-step-two-and-three.py -r results -f data/sequences.fasta -d ALL -t 0.8 -o results/hmm_results -a $UNIPROT_FASTA_DIR -u $UNIPROT_DIR -n $NCBI_DIR --no-step-3
And later step three:
> sbl-FunChaT-step-two-and-three.py --hmm_res_file data/sequences.hmm -a $UNIPROT_FASTA_DIR -u $UNIPROT_DIR -n $NCBI_DIR
Note that the process can be bootstrapped:
> sbl-FunChaT-step-two-and-three.py -r results -f data/sequences.fasta -d D2 -t 0.7 -o results/hmm_results -a $UNIPROT_FASTA_DIR -u $UNIPROT_DIR -n $NCBI_DIR --bootstrap 1 -m
| File Name | Description |
| XML file | An .xml file containing the results of a rigid blocks run between two structures (PDB accession codes: 1OK8 and 1RER). The script should be directed to a folder containing all such comparisons. |
| FASTA file | A fasta file containing all the sequences in fasta format |
| HMM file | A previously computed HMM |
| File Name | Description |
| Plain text file | An .txt dump containing the list of accession codes obtained upon querying UniprotKB with the HMM. |
| Plain text file | A .txt dump containing the list of FILTERED accession codes upon querying UniprotKB with the HMM. |
| Fasta file | A .fasta file containing the sequences used to perform the MSA. |
| Alignment | A .aln file containing the Clustal alignment used to build the HMM. |
| HMM | A file containing the built HMM. |
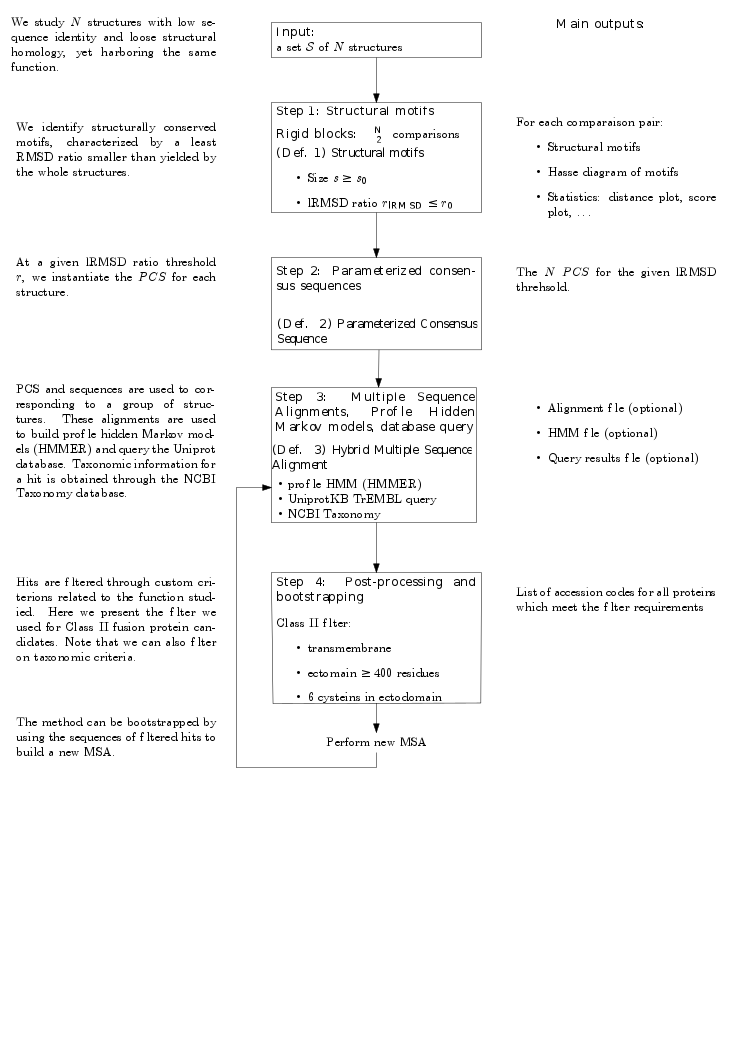 |
| FunChaT: programmer's workflow |
This package uses binaries from the following packages: