 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
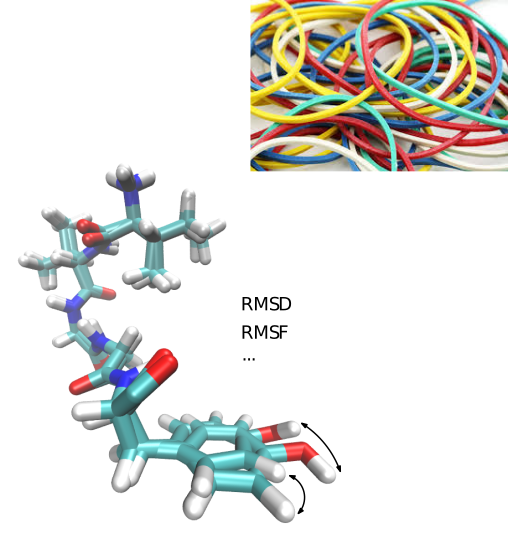
Authors: F. Cazals and R. Tetley
Goals. The root mean square deviation (RMSD) and the least RMSD which are provided in the companion package Molecular_distances are two widely used similarity measures in structural bioinformatics. Yet, they stem from global comparisons, possibly obliterating locally conserved motifs.
To foster our understanding of molecular flexibility, this package, based upon developments presented in [43], provides so-called combined 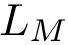
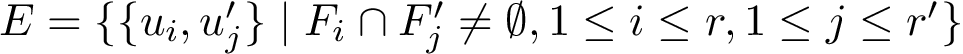
The structural units which may be domains, subdomain, SSE, etc, are defined using the machinery from the package MolecularSystemLabelsTraits
The combined 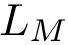
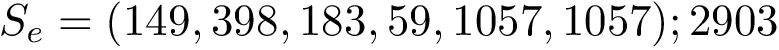
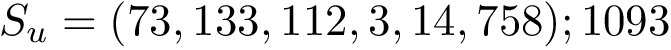
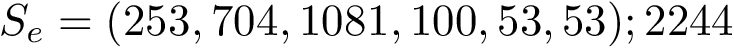
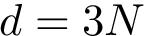
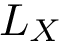
Modes. For the previous programs and as for the package Molecular_distances, four modes are provided:
As noticed above, regions are defined using the labels, as indicated in the package MolecularSystemLabelsTraits.
Two regions with the same label specification may not contain the same number of amino-acids, which typically happens in two cases:
In any case, for each label, a local alignment is run to ensure a 1-1 correspondence between a.a. In the SBL, alignments are defined in the package Alignment_engines.
We actually consider structural alignments in two settings:
We note that for PDB file containing structures of the same polypeptide chain – same SEQRES section, the numberings of amino acids in the ATOMS section are expected to be aligned. See primary-sequences-and-the-pdb-format for details.
The executable corresponding to this case is 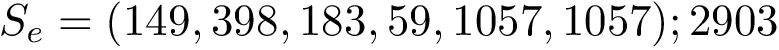
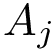
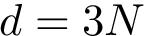
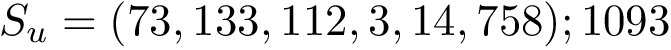
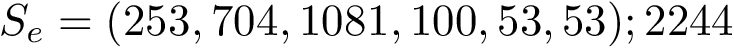
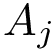
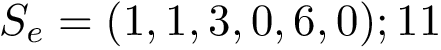
Vertex weighted 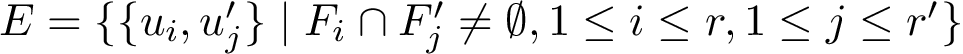
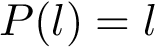
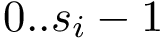
Also consider a set of positive weights 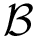
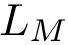
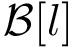
Let 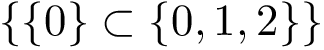
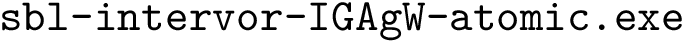
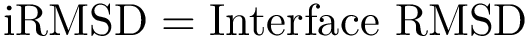
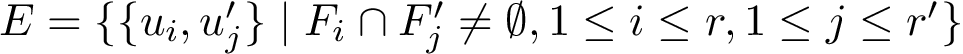
Note that the celebrated 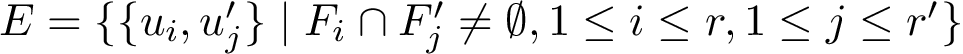
We arrive at the main definition, which combines individual RMSD:
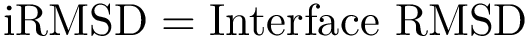
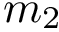
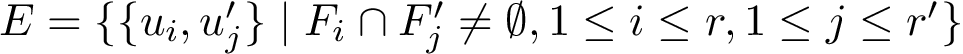
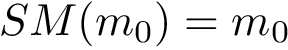
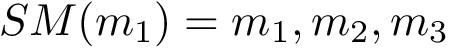
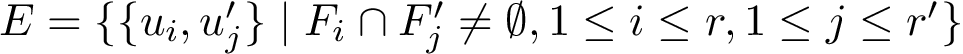
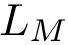
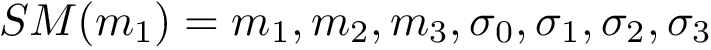
Chain mappings for quaternary structures. Consider the case where one wishes to compare chains from different quaternary structures. In that case, one needs to know the correspondence between the individual chains across these structures. To this end, we define:
Functionalities. The executables 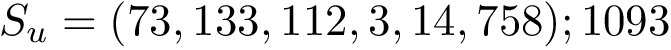
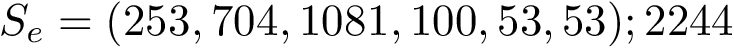
Scenarios. The scenarii discussed below depend on
Options to compare polypeptide chains. The programs 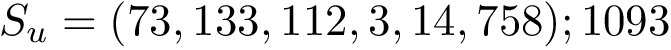
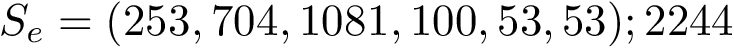
Options to compare conformations. As noticed above, the identity alignment between residues can be used. The options are:
The reader is referred to the jupyter notebook – section Jupyter demo for illustrations.
This case is that of a protein with quaternary structure, in which case we compare the polypeptide chains.
Input.
Output. We report the matrix of 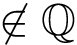
Consider two proteins, with a focus on one chain for each.
Input.
Output. The 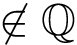
$SBL_DIR/Applications/Molecular_distances_flexible/src/Molecular_distances_flexible/build/sbl-rmsd-flexible-proteins.exe --pdb-file data/pdb_files/SFV-1RER.pdb --domain-labels data/spec_files/SFV.spec --chains A --pdb-file data/pdb_files/TBEV.pdb --domain-labels data/spec_files/TBEV.spec --chains A -d results/twoponec -v -l --allow-incomplete-chains -p 3
Input. Consider two proteins, each of which with n chains, specified as follows:
Output. A matrix of 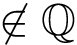
$SBL_DIR/Applications/Molecular_distances_flexible/src/Molecular_distances_flexible/build/sbl-rmsd-flexible-proteins.exe --pdb-file data/pdb_files/SFV-1RER.pdb --domain-labels data/spec_files/SFV.spec --chains ABC --pdb-file data/pdb_files/TBEV.pdb --domain-labels data/spec_files/TBEV.spec --chains ABC -d results/twoponec -v -l --allow-incomplete-chains -p 3
Input.
Output. The matrix of all pairwise 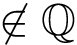
$SBL_DIR/Applications/Molecular_distances_flexible/src/Molecular_distances_flexible/build/sbl-rmsd-flexible-proteins.exe --pdb-file data/pdb_files/SFV-1RER.pdb --domain-labels data/spec_files/SFV.spec --chains A --pdb-file data/pdb_files/TBEV.pdb --domain-labels data/spec_files/TBEV.spec --chains A --pdb-file data/pdb_files/EFF1.pdb --domain-labels data/spec_files/EFF1.spec --chains A -d results/nponec -v -l --allow-incomplete-chains -p 3
Input. Consider now a set of 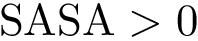
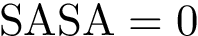
We assume a a mapping between chains is provided – see Def. def-chain-mapping. The corresponding options are the following ones:
Output. We report 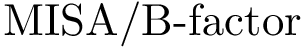
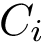
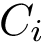
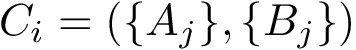
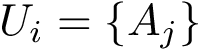
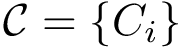
$SBL_DIR/Applications/Molecular_distances_flexible/src/Molecular_distances_flexible/build/sbl-rmsd-flexible-proteins.exe --pdb-file data/pdb_files/SFV-1RER.pdb --domain-labels data/spec_files/SFV.spec --chains ABC --pdb-file data/pdb_files/TBEV.pdb --domain-labels data/spec_files/TBEV.spec --chains ABC --pdb-file data/pdb_files/EFF1.pdb --domain-labels data/spec_files/EFF1.spec --chains ABC --chain-mapping data/mapping.txt -d results/npnc -v -l --allow-incomplete-chains -p 3
Functionalities. The executable 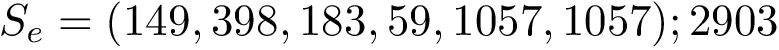
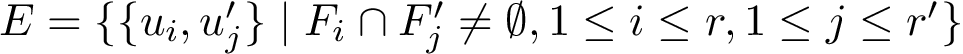
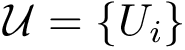
Note that the run scenarios are the same as the previous executable (see Pre-requisites). For example runs, the user is refered to Combined RMSD for proteins .
Structural motifs. As explained in the companion package Structural_motifs , structural motifs are regions showing a structural conservation higher than that of the structures defining them. For two structures, a motif is defined by two sets of a.a. in one-to-one correspondence – that is one set of a.a. on each structure.
Motif graph for overlapping motifs. When several motifs exist for two structures, an important question is to handle them coherently. Since motifs may overlap, we define:
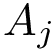
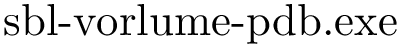
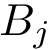
Consider now the case where motifs have been defined for the two structures 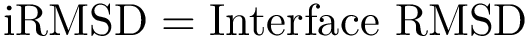
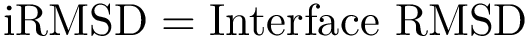
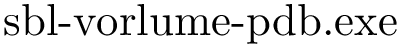
Consider the i-th c.c. of the motif graph. Let 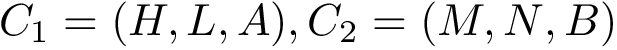
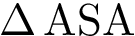
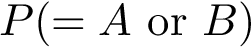
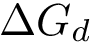
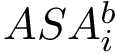
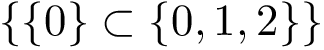
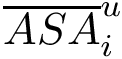
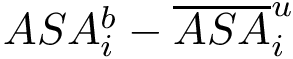
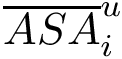
This definition recalled, we note that the 
The input requires two structures (PDB files) and a specification of motifs. The motifs are defined as an identifier followed by a list of aligned residues (example file below).
sbl-rmsd-flexible-motifs.exe --pdb-file data/pdb_files/SFV-1RER.pdb --chains A --pdb-file data/pdb-files/RVFV.pdf --motif-file data/motifs.txt --allow-incomplete-chains -p 3 -d results/motifs -v -l
The implementation rationale behind the three executables is straightforward. Each executable has a workflow consisiting of one loader (SBL::IO::T_Protein_representation_loader, see Protein_representation) and one module. There are two different modules used among the three workflows :
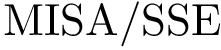
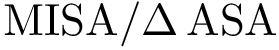
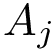
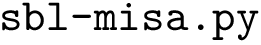
The workflows are extremely basic. One out of the three is displayed below as an example.
T_Local_structural_comparison_workflow:
We note in passing that the implementation of 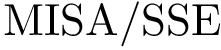
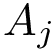
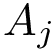
See the following jupyter notebook:
We illustrate calculations involving the so-called combined RMSD or RMSD Comb. As test case, we use class II fusion proteins, decomposing each each polypeptide chain into 23 regions (see preprint by Tetley et al).
import os
import sys
import pdb
from SBL import SBL_pytools
from SBL_pytools import SBL_pytools as sblpyt
odir = "results-new"
sdirs = ["n-pc-one-protein", "two-pc", "n-pc-two-proteins", "n-pc", "m-pc-n-proteins", "motifs"]
for sdir in sdirs:
w = "%s/%s" % (odir, sdir)
if not os.path.exists(w):
os.system( ("mkdir -p %s" % w) )
def cmp_proteins_with_aligner_n_pc_one_protein(odir, aligner, aligner_tag):
osdir = "n-pc-one-protein"
cmd = "%s --pdb-file data/pdb_files/SFV-1RER.pdb --domain-labels data/spec_files/SFV.spec --chains ABC -d %s/%s -v -l" % (aligner,odir,osdir)
print(("Running %s" % cmd))
os.system(cmd)
ofn = "%s/%s/sbl-rmsd-flexible-proteins-%s__weighted_lrmsd.txt" % (odir,osdir,aligner_tag)
odir_osdir = "%s/%s" % (odir,osdir)
file_suffix = "%s__weighted_lrmsd.txt" % aligner_tag
sblpyt.show_text_file(file_suffix, odir_osdir)
aligner_kpax = "sbl-rmsd-flexible-proteins-kpax.exe" # kpax
cmp_proteins_with_aligner_n_pc_one_protein(odir, aligner_kpax, "kpax")
aligner_apurva = "sbl-rmsd-flexible-proteins-apurva.exe" # apurva
cmp_proteins_with_aligner_n_pc_one_protein(odir, aligner_apurva, "apurva")
def cmp_proteins_with_aligner_two_pc(odir, aligner, aligner_tag):
osdir = "two-pc"
cmd = "%s --pdb-file data/pdb_files/SFV-1RER.pdb --domain-labels data/spec_files/SFV.spec --chains A --pdb-file data/pdb_files/TBEV.pdb --domain-labels data/spec_files/TBEV.spec --chains A -d %s/%s -v -l --allow-incomplete-chains -p 3" % (aligner,odir,osdir)
print(("Running %s" % cmd))
os.system(cmd)
odir_osdir = "%s/%s" % (odir,osdir)
file_suffix = "%s__labels_lrmsd.txt" % aligner_tag
sblpyt.show_text_file(file_suffix, odir_osdir)
aligner_kpax = "sbl-rmsd-flexible-proteins-kpax.exe" # kpax
cmp_proteins_with_aligner_two_pc(odir, aligner_kpax, "kpax")
aligner_apurva = "sbl-rmsd-flexible-proteins-apurva.exe" # apurva
cmp_proteins_with_aligner_two_pc(odir, aligner_apurva, "apurva")
def cmp_proteins_with_aligner_n_pc_two_proteins(odir, aligner, aligner_tag):
osdir = "n-pc-two-proteins"
cmd = "%s --pdb-file data/pdb_files/SFV-1RER.pdb --domain-labels data/spec_files/SFV.spec --chains ABC --pdb-file data/pdb_files/TBEV.pdb --domain-labels data/spec_files/TBEV.spec --chains ABC -d %s/%s -v -l --allow-incomplete-chains -p 3" % (aligner,odir,osdir)
print(("Running %s" % cmd))
os.system(cmd)
odir_osdir = "%s/%s" % (odir,osdir)
file_suffix = "%s__weighted_lrmsd.txt" % aligner_tag
sblpyt.show_text_file(file_suffix, odir_osdir)
aligner_kpax = "sbl-rmsd-flexible-proteins-kpax.exe" # kpax
cmp_proteins_with_aligner_n_pc_two_proteins(odir, aligner_kpax, "kpax")
aligner_apurva = "sbl-rmsd-flexible-proteins-apurva.exe" # apurva
cmp_proteins_with_aligner_n_pc_two_proteins(odir, aligner_apurva, "apurva")
def cmp_proteins_with_aligner_n_pc(odir,aligner, aligner_tag):
osdir = "n-pc"
cmd = "%s --pdb-file data/pdb_files/SFV-1RER.pdb --domain-labels data/spec_files/SFV.spec --chains A --pdb-file data/pdb_files/TBEV.pdb --domain-labels data/spec_files/TBEV.spec --chains A --pdb-file data/pdb_files/EFF1.pdb --domain-labels data/spec_files/EFF1.spec --chains A -d %s/%s -v -l --allow-incomplete-chains -p 3" % (aligner,odir,osdir)
print(("Running %s" % cmd))
os.system(cmd)
odir_osdir = "%s/%s" % (odir,osdir)
file_suffix = "%s__weighted_lrmsd.txt" % aligner_tag
sblpyt.show_text_file(file_suffix, odir_osdir)
aligner_kpax = "sbl-rmsd-flexible-proteins-kpax.exe" # kpax
cmp_proteins_with_aligner_n_pc(odir,aligner_kpax, "kpax")
aligner_apurva = "sbl-rmsd-flexible-proteins-apurva.exe" # apurva
cmp_proteins_with_aligner_n_pc(odir,aligner_apurva, "apurva")
def cmp_proteins_with_aligner_m_pc_n_chains(odir,aligner, aligner_tag):
osdir = "m-pc-n-proteins"
cmd = "%s --pdb-file data/pdb_files/SFV-1RER.pdb --domain-labels data/spec_files/SFV.spec --chains ABC --pdb-file data/pdb_files/TBEV.pdb --domain-labels data/spec_files/TBEV.spec --chains ABC --pdb-file data/pdb_files/EFF1.pdb --domain-labels data/spec_files/EFF1.spec --chains ABC --chain-mapping data/mapping.txt -d %s/%s -v -l --allow-incomplete-chains -p 3" % (aligner,odir,osdir)
print(("Running %s" % cmd))
os.system(cmd)
odir_osdir = "%s/%s" % (odir,osdir)
file_suffix = "%s__weighted_lrmsd_chain_0.txt" % aligner_tag
sblpyt.show_text_file(file_suffix, odir_osdir)
file_suffix = "%s__weighted_lrmsd_chain_1.txt" % aligner_tag
sblpyt.show_text_file(file_suffix, odir_osdir)
file_suffix = "%s__weighted_lrmsd_chain_2.txt" % aligner_tag
sblpyt.show_text_file(file_suffix, odir_osdir)
aligner_kpax = "sbl-rmsd-flexible-proteins-kpax.exe" # kpax
cmp_proteins_with_aligner_m_pc_n_chains(odir,aligner_kpax, "kpax")
aligner_apurva = "sbl-rmsd-flexible-proteins-apurva.exe" # apurva
cmp_proteins_with_aligner_m_pc_n_chains(odir,aligner_apurva, "apurva")
# cmp proteins using motifs
#i################################################################################
def cmp_proteins_with_motifs():
## 4.2 Combined RMSD for structural motifs
osdir = "motifs"
aligner = "sbl-rmsd-flexible-motifs.exe"
cmd = "%s --pdb-file data/pdb_files/SFV-1RER.pdb --chains A --pdb-file data/pdb_files/RVFV.pdb --motif-file data/motifs.txt --allow-incomplete-chains -p 3 -d %s/%s -v -l" % (aligner,odir,osdir)
print(("Running %s" % cmd))
os.system(cmd)
cmp_proteins_with_motifs()