 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
Authors: F. Cazals and R. Tetley
The 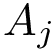
In short, having computed an initial 3D superposition of the two structures based on so-called seeds, Kpax performs a number of iterations involving two steps [165] :
To get insights on the similarity measure used, we only recall the definition of the score 
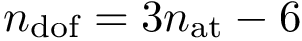
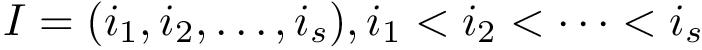
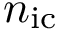

With these, the total Kpax score for two residues of the two structures to be aligned reads as
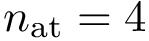
The main input consists of two PDB/mmCIF files.
Example call:
sbl-kpax-iterative-alignment.exe -f data/1anx.pdb -f data/2ran.pdb --load-chains A --load-chains A -d output-directory --log
The output consists of the following files:
A 1 A 1 A 2 A 2 A 4 A 3 A 5 A 4
The SBL provides VMD, PyMOL, and Web plugins for sbl-kpax-iterative-alignment.exe. Launch the VMD plugin from the SBL catalog under the Extensions menu, start the PyMOL plugin by running sbl_kpax in the PyMOL command line, and open the Web plugin from the SBL web plugins launcher page (served locally via sbl-web-plugins-launcher). All three plugins expose the same GUI and capabilities.
 |
sbl-kpax plugin user interface. The user interface for the sbl-kpax plugin. The typical workflow is: Step 1: Load two molecular structures from PDB files. Step 2: Specify the chains to be aligned from each structure. Step 3: Choose an output directory (optional, a default is provided). Step 4: Launch the computation. Step 5: Inspect the results in the viewer. Step 6: Review the calculation statistics, alignment strings, and alignment graph in the GUI. |
The Kpax implementation is non trivial, as it requires manipulating sequences, structures, and alignments. Part of the complexity comes from the fact that the classes used for Kpax are also used to compute molecular distances (package Molecular_distances_flexible) and search structural motifs (package Structural_motifs).
The overall architecture is as follows:
More specifically:
Let us now detail these components.
Consider the problem of aligning two structures structure_1 and structure_2, respectively containing 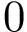
Structures.
As explained below, T_Structure_for_kpax is used to instantiate SBL::CSB::T_Iterative_aligner<class StructureType, class SeederType, class ScoreComputer>
Constructing structures from PDB/mmCIF files. To load a PDB/mmCIF files, one iterates on all chains of a protein, collects the 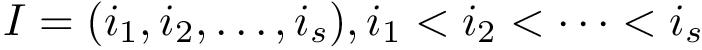
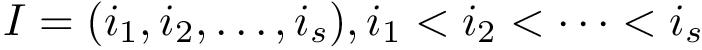
The resulting data structure is an instance of T_Structure_for_kpax, which, as explained above, is a vector of T_Alignment_residue.
Two comments are in order:
![$R_1=[0, n_1-1]$](form_687.png)
![$R_2=[0, n_2-1]$](form_688.png)
Dumping alignments. As just explained, an alignment is recorded as a list of aligned residues 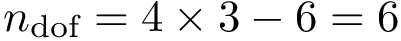
On the other hand, the alignment structure is a vector of alignment residues, such that each contains its original residue.
Therefore, the refs of each aligned residue are easily retrieved from the original residue. e.g., the resid (protein sequence ids) are directly as structure_1.unit_id() or structure_1.residue_sequence_number() .
Iterative_alignment : a generic framework for iterative aligners, providing in particular the generic implementation of Kpax. Provides in particular the classes SBL::CSB::T_Iterative_aligner and SBL::CSB::T_Kpax .
The engine level combines the algorithm (aligner) and all the calculation of all statistics of interest. Note in particular the following inheritances hierarchy:
SBL::CSB::T_Alignment_engine_structures_kpax inherits from SBL::CSB::T_Alignment_engine_structures inherits from SBL::CSB::T_Alignment_engine
Importantly, the class SBL::CSB::T_Alignment_engine has two data members of type StructureType, named m_sos_1 and m_sos_2 (for my sequence_or_structure).
The traits class used to instantiate Kpax is provided by the package Kpax itself. It defines types that may be ascribed to two groups:
typedef SBL::CSB::T_Alignment_engine_structures_kpax<Structure> Alignment_engine;
An application in the SBL is graph connecting modules, which inherit from the base class Module_base .
Since Kpax addresses an alignment problem, the generic workflow used is SBL::CSB::T_Iterative_alignment_workflow .
The module for Kpax is SBL::CSB::T_Alignment_structures_module, which implements the functions
See the following jupyter demo:
The main options of the align method in the next cell are:
import re #regular expressions
import sys #misc system
import os
import pdb
import shutil # python 3 only
def align(pdb1, pdb2, chains1 = None, chains2 = None):
odir = "tmp-results"
if os.path.exists(odir):
os.system("rm -rf %s" % odir)
os.system( ("mkdir %s" % odir) )
# check executable exists and is visible
exe = shutil.which("sbl-kpax-iterative-alignment.exe")
if exe:
print(("Using executable %s\n" % exe))
cmd = "%s -f %s -f %s -v -l -d %s" %\
(exe, pdb1, pdb2, odir)
if chains1 != None:
cmd += " --chains %s" % chains1
if chains2 != None:
cmd += " --chains %s" % chains2
print(cmd)
os.system(cmd)
cmd = "ls %s" % odir
ofnames = os.popen(cmd).readlines()
print("All output files:",ofnames)
#find the log file and display log file
cmd = "find %s -name *log.txt" % odir
lines = os.popen(cmd).readlines()
if len(lines) > 0:
lfname = lines[0].rstrip()
print("Log file is:", lfname)
log = open(lfname).readlines()
for line in log: print(line.rstrip())
else:
print("Executable not found")
# Simple example with 2 helices
print("Marker : Calculation Started")
align("data/theor-helix-5.pdb","data/theor-helix-5.pdb")
print("Marker : Calculation Ended")
Marker : Calculation Started Using executable /user/fcazals/home/projects/proj-soft/sbl-install/bin/sbl-kpax-iterative-alignment.exe /user/fcazals/home/projects/proj-soft/sbl-install/bin/sbl-kpax-iterative-alignment.exe -f data/theor-helix-5.pdb -f data/theor-helix-5.pdb -v -l -d tmp-results All output files: ['sbl-kpax__alignment.dot\n', 'sbl-kpax__alignment.txt\n', 'sbl-kpax__log.txt\n'] Log file is: tmp-results/sbl-kpax__log.txt Running: /user/fcazals/home/projects/proj-soft/sbl-install/bin/sbl-kpax-iterative-alignment.exe -f data/theor-helix-5.pdb -f data/theor-helix-5.pdb -v -l -d tmp-results Protein representation loader Statistics: Number of loaded PDB files: 2 Details for each file: -- file 1 data/theor-helix-5.pdb: -- -- File exists: Yes -- -- File is valid: Yes -- -- Number of models: 1 -- -- Number of loaded atoms (of model with lowest index): 25 -- -- Number of loaded SS bonds: 0 -- -- Number of discarded atoms based on model id (all models): 0 -- -- Number of discarded atoms based on chain id (all models): 0 -- -- Number of discarded atoms based on b factor beeing too high (all models): 0 -- -- Number of discarded waters (all models): 0 -- -- Number of discarded hetatoms (all models): 0 -- -- Number of discarded hydrogens (all models): 0 -- -- Number of discarded alternate location atoms (all models): 0 -- -- Number of actually discarded atoms (all models): 0 -- file 2 data/theor-helix-5.pdb: -- -- File exists: Yes -- -- File is valid: Yes -- -- Number of models: 1 -- -- Number of loaded atoms (of model with lowest index): 25 -- -- Number of loaded SS bonds: 0 -- -- Number of discarded atoms based on model id (all models): 0 -- -- Number of discarded atoms based on chain id (all models): 0 -- -- Number of discarded atoms based on b factor beeing too high (all models): 0 -- -- Number of discarded waters (all models): 0 -- -- Number of discarded hetatoms (all models): 0 -- -- Number of discarded hydrogens (all models): 0 -- -- Number of discarded alternate location atoms (all models): 0 -- -- Number of actually discarded atoms (all models): 0 Covalent Structure File Loader statistics: -- -- Number of molecular covalent structures: 2 -- Molecular covalent structure 0: -- -- Number of particles: 52 -- -- Number of embedded particles: 25 -- -- Number of bonds: 51 -- -- Number of embedded bonds: 24 -- -- Number of disulfide bonds: 0 -- Molecular covalent structure 1: -- -- Number of particles: 52 -- -- Number of embedded particles: 25 -- -- Number of bonds: 51 -- -- Number of embedded bonds: 24 -- -- Number of disulfide bonds: 0 Protein Representation Loader statistics: Number of loaded proteins: 2 Details for each protein : -- structure 1: -- -- Number of chains: 1 -- -- Number of residues: 5 -- -- Number of particles: 52 -- -- Number of embedded particles: 25 -- -- Number of bonds: 51 -- -- Number of embedded bonds: 24 -- structure 2: -- -- Number of chains: 1 -- -- Number of residues: 5 -- -- Number of particles: 52 -- -- Number of embedded particles: 25 -- -- Number of bonds: 51 -- -- Number of embedded bonds: 24 Iterative alignment Starting module SBL::Modules::T_Alignment_structures_module... +++Seeder score: 5.000000 +++Performing iterative alignment with 1 seeds... (non id case)Iter 1 out of 1 with seed of size 5 (i, lRMSD, size) (0, 0.00000, 4) Converged to a 0-cycle after 2 iterations Decreasing lRMSDs true Statistics: Alignment length: 4 Alignment score : 4.000000 Alignment identity percentage: 1.000000 Alignment similarity percentage: 1.000000 Alignment lRMSD : 0.000000 Alignment dRMSD : 0.000000 Report... Ending module SBL::Modules::T_Alignment_structures_module... End Run General Statistics: Times elapsed for computations (in seconds): -- Iterative alignment: 0.001094 Total: 0.001094 Marker : Calculation Ended
Seeder_DP_score:alignment size:5
# Example from the molecular motions DB, see http://www0.molmovdb.org/cgi-bin/motion.cgi?ID=anxtrp
# Motion in Annexin V (Trp motion) [anxtrp]
# 1ANX, 1AVR, 2RAN
print("Marker : Calculation Started")
#align("data/theor-helix-5.pdb","data/theor-helix-5.pdb")
align("data/1anx.pdb","data/2ran.pdb")
print("Marker : Calculation Ended")
Marker : Calculation Started Using executable /user/fcazals/home/projects/proj-soft/sbl-install/bin/sbl-kpax-iterative-alignment.exe /user/fcazals/home/projects/proj-soft/sbl-install/bin/sbl-kpax-iterative-alignment.exe -f data/1anx.pdb -f data/2ran.pdb -v -l -d tmp-results
Seeder_DP_score:alignment size:326
All output files: ['sbl-kpax__alignment.dot\n', 'sbl-kpax__alignment.txt\n', 'sbl-kpax__log.txt\n'] Log file is: tmp-results/sbl-kpax__log.txt Running: /user/fcazals/home/projects/proj-soft/sbl-install/bin/sbl-kpax-iterative-alignment.exe -f data/1anx.pdb -f data/2ran.pdb -v -l -d tmp-results Protein representation loader Statistics: Number of loaded PDB files: 2 Details for each file: -- file 1 data/1anx.pdb: -- -- File exists: Yes -- -- File is valid: Yes -- -- Number of models: 1 -- -- Number of loaded atoms (of model with lowest index): 7437 -- -- Number of loaded SS bonds: 0 -- -- Number of discarded atoms based on model id (all models): 0 -- -- Number of discarded atoms based on chain id (all models): 0 -- -- Number of discarded atoms based on b factor beeing too high (all models): 0 -- -- Number of discarded waters (all models): 418 -- -- Number of discarded hetatoms (all models): 445 -- -- Number of discarded hydrogens (all models): 0 -- -- Number of discarded alternate location atoms (all models): 91 -- -- Number of actually discarded atoms (all models): 445 -- file 2 data/2ran.pdb: -- -- File exists: Yes -- -- File is valid: Yes -- -- Number of models: 1 -- -- Number of loaded atoms (of model with lowest index): 2487 -- -- Number of loaded SS bonds: 0 -- -- Number of discarded atoms based on model id (all models): 0 -- -- Number of discarded atoms based on chain id (all models): 0 -- -- Number of discarded atoms based on b factor beeing too high (all models): 0 -- -- Number of discarded waters (all models): 108 -- -- Number of discarded hetatoms (all models): 125 -- -- Number of discarded hydrogens (all models): 573 -- -- Number of discarded alternate location atoms (all models): 0 -- -- Number of actually discarded atoms (all models): 698 Covalent Structure File Loader statistics: -- -- Number of molecular covalent structures: 2 -- Molecular covalent structure 0: -- -- Number of particles: 15324 -- -- Number of embedded particles: 7437 -- -- Number of bonds: 15417 -- -- Number of embedded bonds: 7530 -- -- Number of disulfide bonds: 0 -- Molecular covalent structure 1: -- -- Number of particles: 5052 -- -- Number of embedded particles: 2487 -- -- Number of bonds: 5081 -- -- Number of embedded bonds: 2516 -- -- Number of disulfide bonds: 0 Protein Representation Loader statistics: Number of loaded proteins: 2 Details for each protein : -- structure 1: -- -- Number of chains: 3 -- -- Number of residues: 948 -- -- Number of particles: 15324 -- -- Number of embedded particles: 7437 -- -- Number of bonds: 15417 -- -- Number of embedded bonds: 7530 -- structure 2: -- -- Number of chains: 1 -- -- Number of residues: 316 -- -- Number of particles: 5052 -- -- Number of embedded particles: 2487 -- -- Number of bonds: 5081 -- -- Number of embedded bonds: 2516 Iterative alignment Starting module SBL::Modules::T_Alignment_structures_module... +++Seeder score: 286.733085 +++Performing iterative alignment with 2 seeds... (non id case)Iter 1 out of 2 with seed of size 312 (i, lRMSD, size) (0, 0.53412, 315) Converged to a 0-cycle after 2 iterations Decreasing lRMSDs true (non id case)Iter 2 out of 2 with seed of size 316 (i, lRMSD, size) (0, 0.53412, 315) Converged to a 0-cycle after 2 iterations Decreasing lRMSDs true Statistics: Alignment length: 315 Alignment score : 309.044049 Alignment identity percentage: 0.917460 Alignment similarity percentage: 0.955556 Alignment lRMSD : 0.534116 Alignment dRMSD : 0.449635 Report... Ending module SBL::Modules::T_Alignment_structures_module... End Run General Statistics: Times elapsed for computations (in seconds): -- Iterative alignment: 7.027162 Total: 7.027162 Marker : Calculation Ended