 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
Packages | |
| Alignment_engines | |
 | Engines performing alignments of sequences or structures Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus and S. Marillet and R. Tetley |
| Alpha_complex_of_molecular_model | |
 | 3D weighted |
| Apurva | |
 | Structural alignment ... Reference Manual – User Manual . Authors: N. Malod-Dognin and R. Andonov |
| Biomolecule_representation | |
 | Here you should put an abstract of your package Reference Manual – User Manual . Authors: F. Cazals and C. Robert |
| Buried_surface_area | |
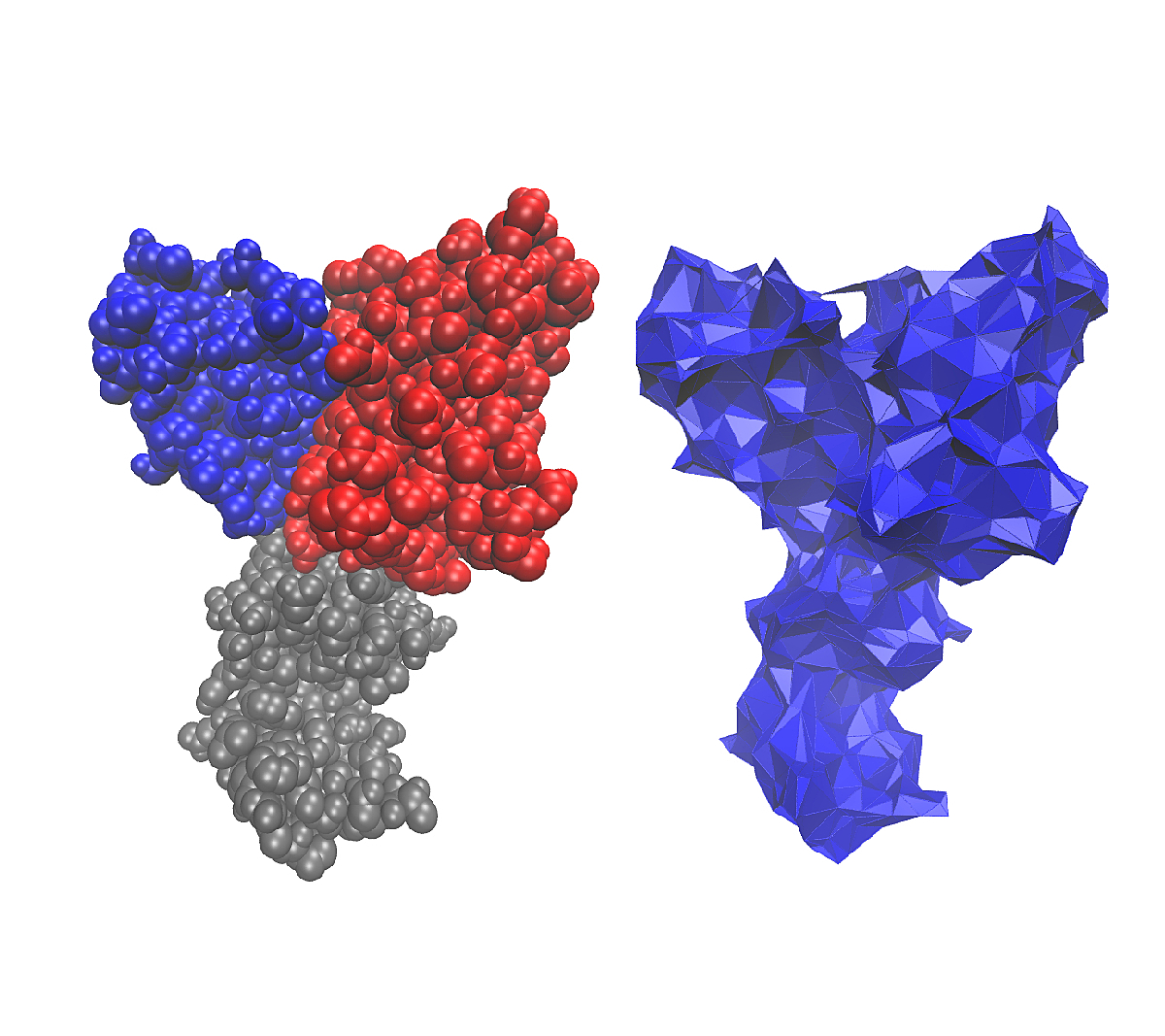
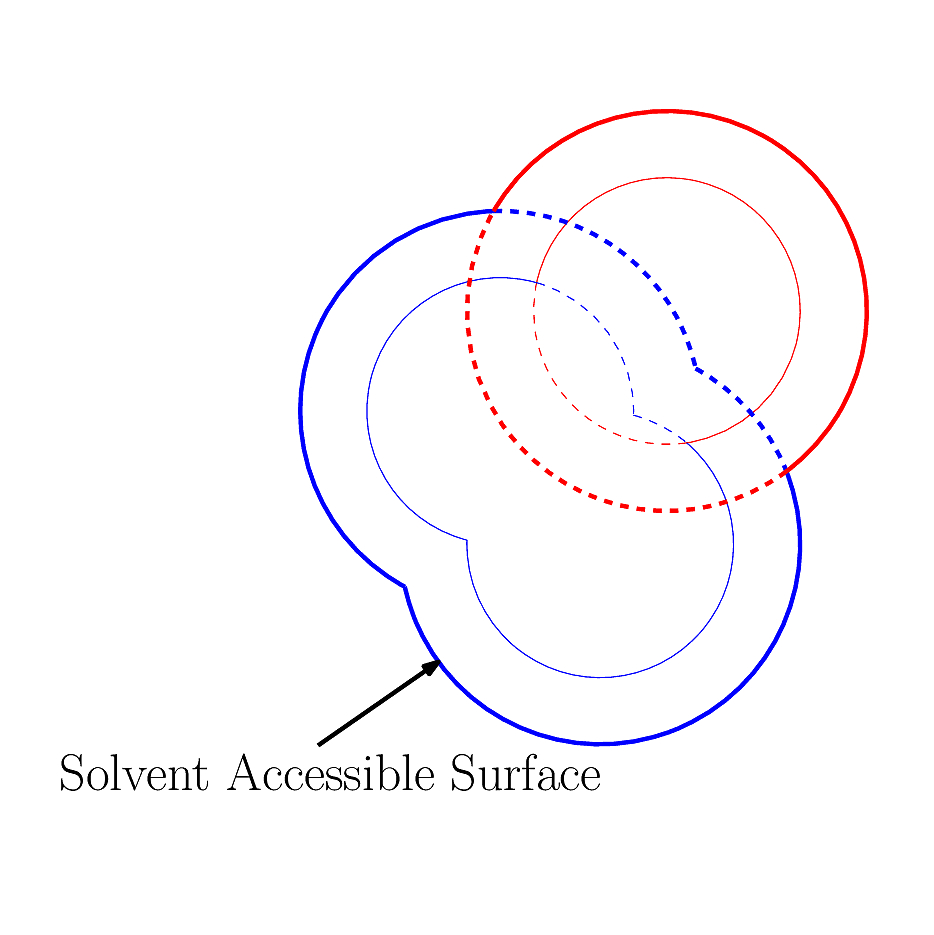 | Algorithm for computing the surface area of a molecule buried in another molecule Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus |
| DB_manipulator | |
 | A package providing methods to ease the access to various protein related databases Reference Manual – User Manual . Authors: R. Tetley and F. Cazals |
| HMMER_Wrapper | |
 | A python wrapper for HMMER Reference Manual – User Manual . Authors: R. Tetley and F. Cazals |
| Iterative_alignment | |
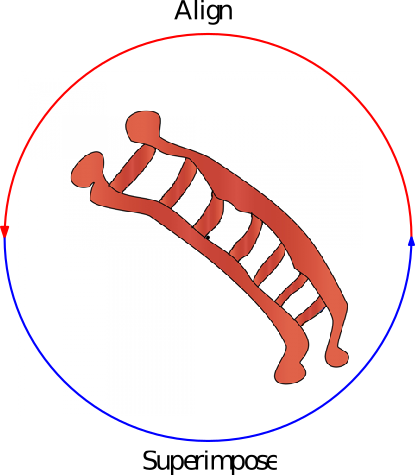 | A generic algorithm performing rounds of dynamic programming alignment and rigid superposition until a final structural alignment is reached. Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus and R. Tetley |
| Linear_polymer_representation | |
 | Generic implementation for linear polymers Reference Manual – User Manual . Authors: G. Carrière and F. Cazals and C. Robert |
| Molecular_conformation | |
 | A package providing classes to store biomolecule conformations. Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus and C. Le Breton |
| Molecular_coordinates | |
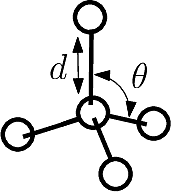 | Conversion between coordinate systems for a molecule Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus and T. O'Donnell |
| Molecular_covalent_structure | |
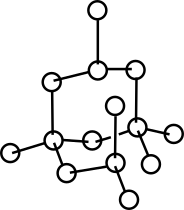 | Representation of the topology of a molecule Reference Manual – User Manual . Authors: F. Cazals and A. Chevallier and T. Dreyfus |
| Molecular_distances | |
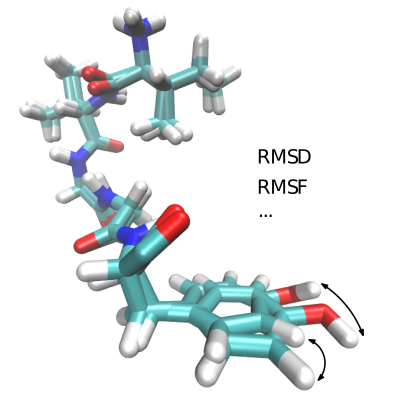 | Algorithms computing different kind of distances between molecular conformations. Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus |
| Molecular_geometric_model_classifier | |
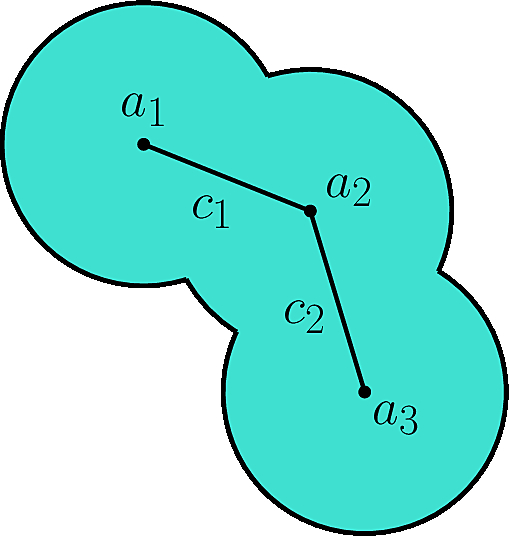 | Data structure classifying the contacts between atoms of a molecular geometric model. Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus |
| Molecular_interfaces | |
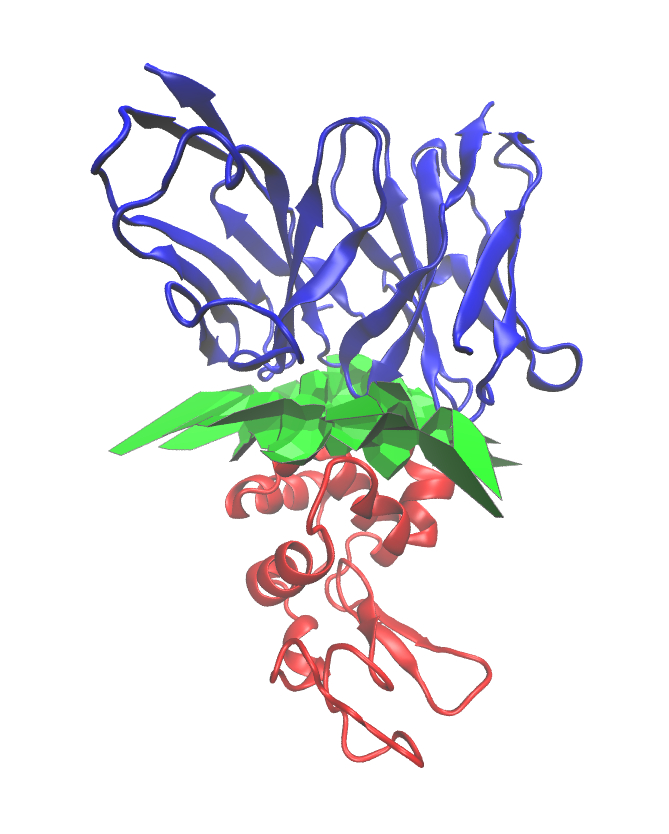 | Data structures and algorithms for computing the interface patches between different partners in a molecule Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus |
| Molecular_potential_energy | |
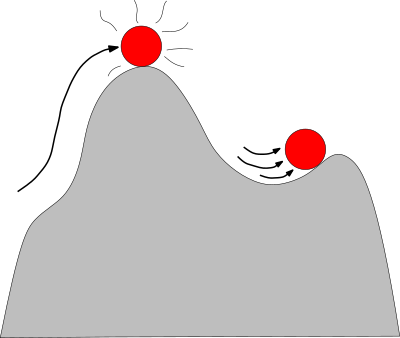 | Computation of the potential energy of a molecule Reference Manual – User Manual . Authors: F. Cazals and A. Chevallier and T. Dreyfus |
| Molecular_structure_classifier | |
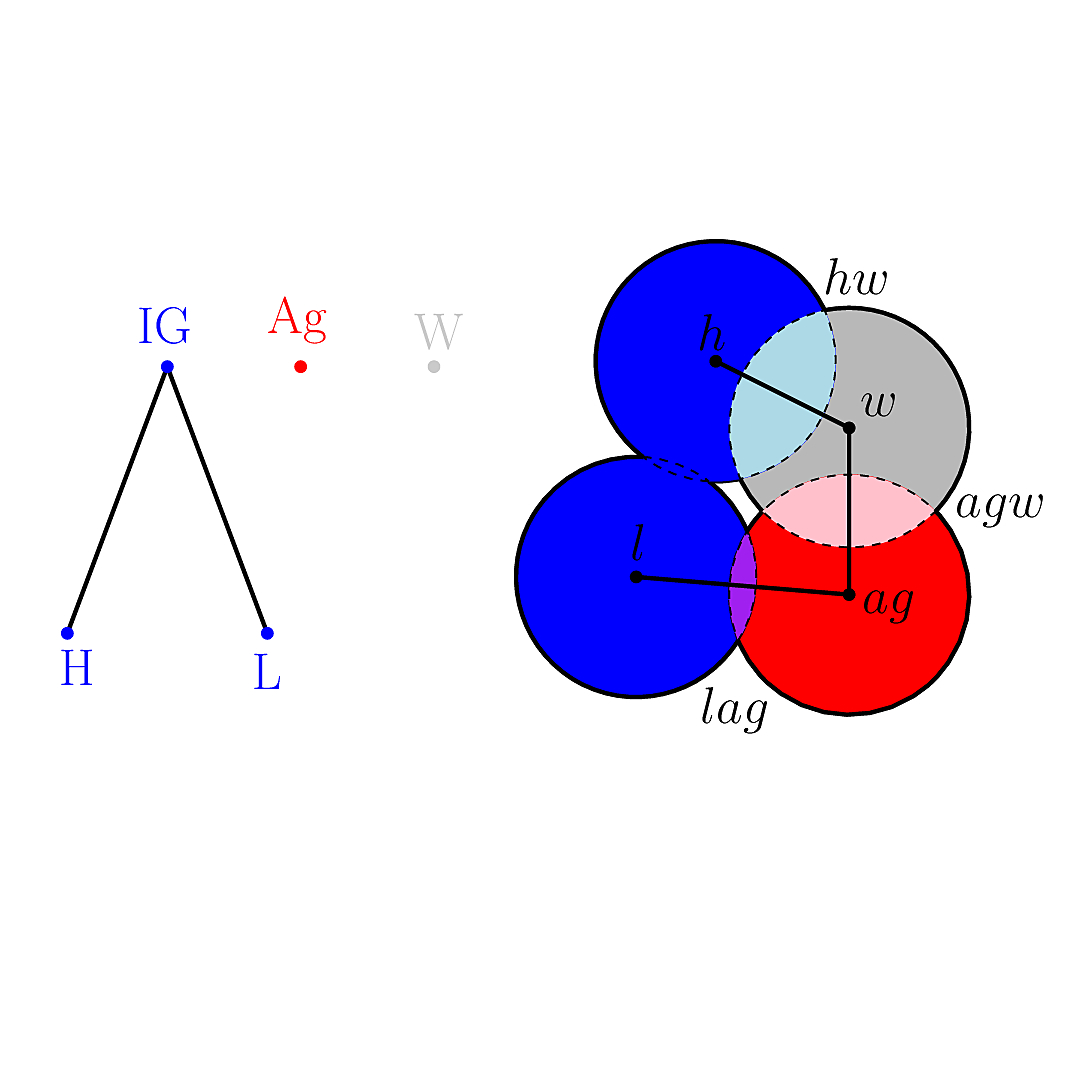 | Data structure classifying contacts between atoms of different partners in a molecule Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus |
| Molecular_system | |
 | A package providing classes to store and architecture molecular data in a structured manner. Reference Manual – User Manual . Authors: F. Cazals and S. Loriot and C. Le Breton |
| Nucleic_acid_representation | |
 | A package providing classes to easily access relevant information attached to Nucleic Acid representations in the SBL. Reference Manual – User Manual . Authors: F. Cazals and C. Robert |
| Pointwise_interactions | |
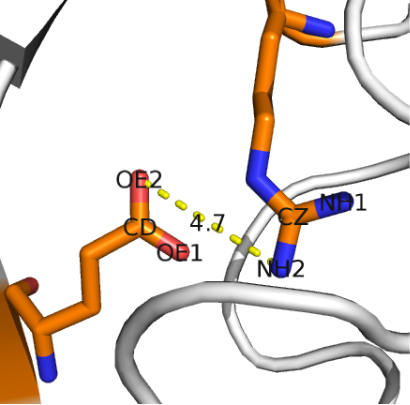 | A package to report pointwise interactions from atomic data Reference Manual – User Manual . Authors: F. Cazals and R. Tetley |
| Protein_representation | |
 | A package providing classes to easily access all pieces of information attached to proteins. Reference Manual – User Manual . Authors: F. Cazals and G. Carrière and T. Dreyfus and C. Robert and R. Tetley |
| Protein_sequence_annotator | |
 | A package providing methods to annotate protein sequences Reference Manual – User Manual . Authors: R. Tetley and F. Cazals |
| Real_value_function_minimizer | |
 | Under Construction Reference Manual – User Manual . Authors: A. Roth and F. Cazals and T. Dreyfus |
| Tertiary_quaternary_structure_annotator | |
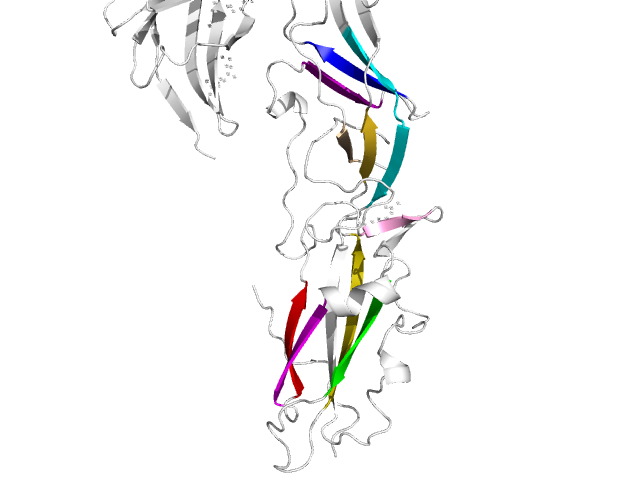 | A package to annotate contacts within tertiary and quaternary structures Reference Manual – User Manual . Authors: F. Cazals and and T. Dreyfus and R. Tetley |
| Transition_graph_traits | |
 | Under Construction Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus |
| Tripeptide_loop_closure | |
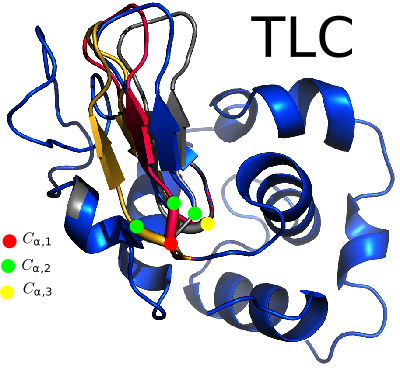 | Here you should put an abstract of your package Reference Manual – User Manual . Authors: F. Cazals and T.O'Donnell |
Computational Structural Biology.
This set of packages present specific algorithms used in Computational Structural Biology, such as the classification of molecular structures, the definition of molecular interfaces, and the computation of buried surface areas.