 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
The Structural Bioinformatics Library (SBL) is a generic C++/python library providing combinatorial, geometric and topological tools to solve problems in computational structural biology. It ambitions to provide high level concepts and methods to foster our understanding of the relationship between the structure and the function of macro-molecules and their complexes,
The SBL runs on most Unix systems (Linux and MacOS), and also on Windows.
Its design is meant to accommodate both the variety of molecular geometric models coding the physical and chemical properties of macro-molecular structures, and the variety of operations undertaken on these models. The models supported either consist of unions of balls (e.g., van der Waals models), or representations based on Cartesian or internal coordinates. The operations deal with complementary aspects, namely geometric, topological, and combinatorial aspects. Software development in this context is especially challenging due to the interactions between these complex models and operations.
Main software components:
| Applications | An application solves a specific biophysical problem |
| Core | Low-level generic C++ classes |
| Models | C++ models matching bio-physical C++ concepts |
| Modules | Black boxes used in applications |
Resources:
| Guides | Documentation Guide | Installation Guide | |
| Tutos | End-User: Tutorial | Developers Using SBL: Tutorial | Contributors : Manual |
| Others | Terminology and Concepts | Bibliography | Data Models |
Overview of the SBL
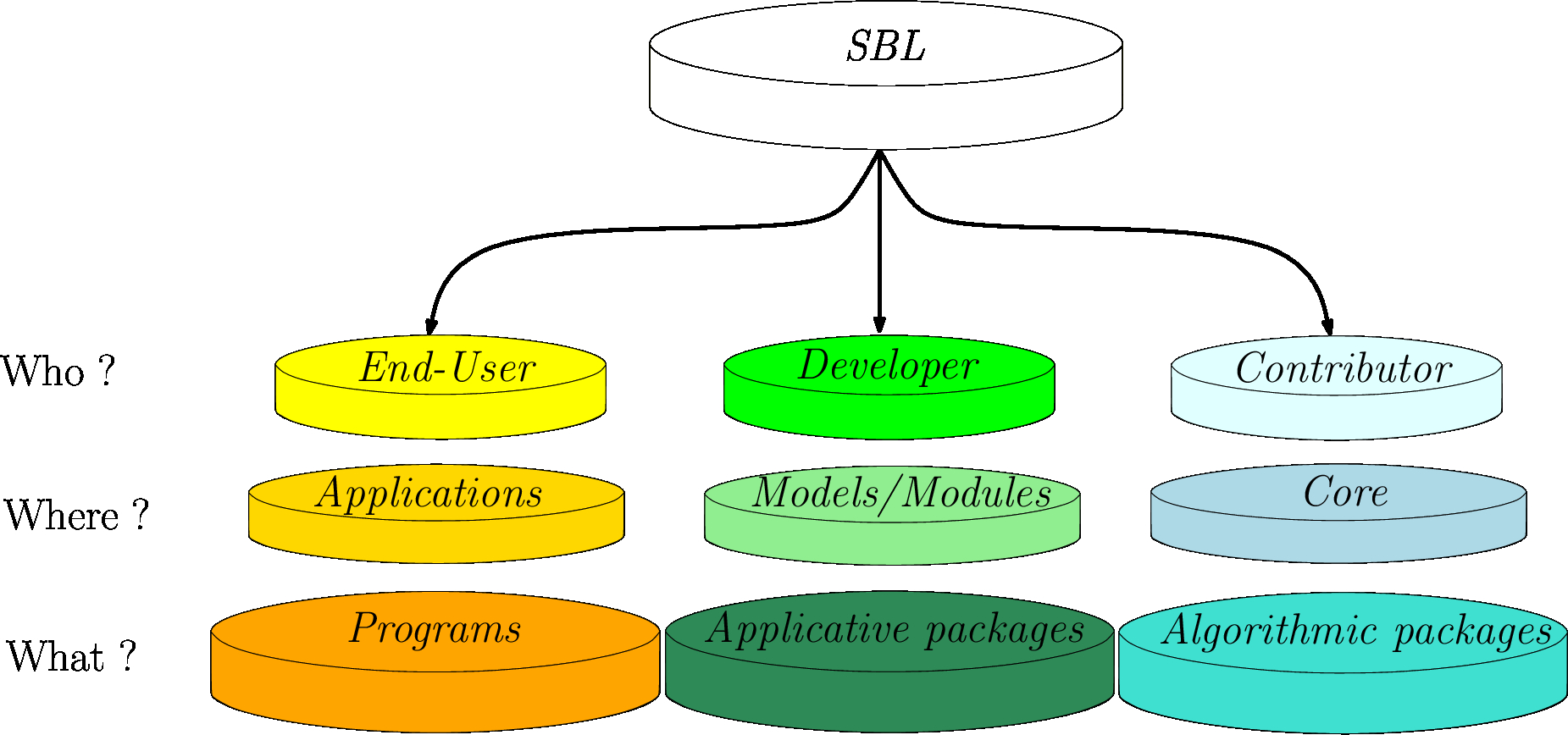 |
| Focus areas for end-users, developers, and contributors. |
Publications.
For a list of publications describing or using software components from the SBL, see Publications .
Applications have been defined above. An iconic example is the calculation of molecular surfaces and volumes–the input may indeed consist of a collection of balls which can be atoms or pseudo-atoms of any kind, see Space_filling_model_surface_volume.
Applications from the SBL are ascribed to the following work packages:
| Space Filling Model | Analyzing molecular geometric models defined by union of balls |
| Conformational Analysis | Modeling molecular flexibility and energy landscapes |
| Assemblies | Modeling macro-molecular assemblies involving from tens to hundreds of macro-molecules |
| Integrated Analysis | Solving problems requiring various building blocks / applications |
| Data Management | Automating data storage, parsing and retrieval |
| Data Analysis | Data analysis and statistical processing |
At the end-user level, the SBL provides binary programs that can be run from the command-line. To ease the process of using them, the SBL also provides plugins for the most popular molecular visualization tools:
The installation of these plugins is automated, as explained on the Installation guide. The installation script is available on the SBL applications page. In a nutshell, the install scripts performs the following tasks:
downloads the library and the pre-compiled programs,
Updating the plugins is done in the exact same way—the scripts detect whether the library and the plugins are already installed.
Once installed, the programs from the SBL offer two types of functionalities:
Plugins, each associated with a GUI. A plugin calls an executable, which typically generates graphical outputs directly sent to the 3D viewer.
These functionalities are described in the User Manual of the SBL applications concerned.
Development with the SBL can occur at two levels.
Low level developments. C++ developments may use classes from Core and Models. In fact, such developments are equivalent to those based upon C++ libraries such as CGAL or boost C++ libraries . It should be noticed that the SBL heavily relies on these libraries. The Core is organized into five sub-sections:
| CADS | Combinatorial Algorithms and Data Structures |
| GT | Geometry and Topology |
| CSB | Computational Structural Biology |
| IO | Input / Output |
| Modules | Modules to define workflows |
These packages implement algorithms not available elsewhere, or which available in non-generic guise. Due to the modular structure of the library, should valuable implementations be made available outside the SBL (e.g. in CGAL or boost), a substitution may occur.
Intermediate level developments. To avoid the intricacies inherent to low level developments, one should use modules. Modules indeed allow the development of applications without the burden of instantiating low level classes. In fact, once modules are available, designing an application merely consists of connecting modules.
The SBL is interoperable with existing molecular modeling systems, at several levels:
At the library level, our state-of-the-art algorithms (e.g. the computation of molecular surfaces and volumes) can be integrated within existing software (e.g. molecular dynamics software), by instantiating the required classes from Core, or using the adequate modules.
At the application level, our applications can easily be integrated within processing pipelines, since the format used for input and output are standard ones. (For input, the PDB format can always be used. For output, our applications generate XML files.)
The SBL is released under a proprietary open source licence. In a nutshell, academic users can use and modify the code at their discretion, for private purposes. But distributing these changes or doing business is forbidden. However, novel capabilities matching the design choices of the library will be welcome.
The version of SBL is made of three numbers (see the Software versioning wikipedia page for further details):
The source code is distributed from a tarball or a git repository from the download page of the SBL web-site.
Bugzilla is used to handle user's feedback and bug tracking.