 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|

Authors: R. Tetley and F. Cazals
Profiling families of proteins through sequence information has been at the heart of bio-informatics. Of particular interest are hidden Markov models, which, upon processing a multiple sequence alignment, builds a probablistic model enabling large database queries. This package is simply a python wrapper for HMMER (http://hmmer.org/), a software suite which, among other things, allows to build HMM and query databases from a built HMM.
To build an HMM, a user is expected to input a multiple sequence alignment of the scrutinized proteins. This package uses the Clustal Omega algorithm to compute such an alignment (http://www.clustal.org/omega/).
Profile hidden Markov models are specific Markov models meant to generate protein sequences, see [120] [86] as well as general sources on HMM .
Given a HMM and a target sequence, two questions are easily answered using dynamic programming.
Phrased differently, the Viterbi algorithm identifies the optimal alignment 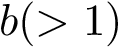

The aforementioned scores can be used to retrieve the sequences within a database in best agreement with the model. Assessing the significance of a score is best done via a so-called e-value (expectation value, as opposed to p-value), which gives an estimate of the number of sequences that would get this score (or better) in a database of the same size containing only random sequences [156] [118] [142].
Intuitively, the p-value and the e-value are related by 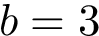
The e-value calculation requires the distribution of scores for such random sequences, so that the exact formula depends on the alignment / scoring method and statistical properties thereof, see [142] and [106] .
This package provides a single python module (SBL/HMMER_Wrapper.py). The SBL::HMMER_Wrapper::HMM class expects as input a signature (the name of the HMM, generally built by combining the names of the proteins used to build it), a dictionnary with protein names as keys and their FASTA sequences as values and the path to a FASTA data base (to launch the query). An additional output path is required should the user whish to save the files generated by the different programs.
Upon running an HMM object, the program will build temporary files containing the fasta sequences. The program will then proceed to run Clustal Omega, then hmmbuild and hmmsearch. All the results are parsed.
The user can access results through the get_results function, which requires a threshold. The accession codes with e-values which are smaller than the provided threshold are returned.
Below is a simple example in which a HMM is instantiated from sequences contained in a file. Only the results below the 0.1 threshold are printed.
In order to run, the user needs to have the following programs installed: