 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
Tutorial guiding an end-user for using the SBL programs.
This tutorial aims to explain how to use the programs developed within SBL. It is organized as follows:
Note that it is assumed that the SBL library was installed as specified in the Installation Guide.
In the following, we describe where are located the SBL programs and the basic options that are common to all the SBL programs.
If the cmake variable SBL_APPLICATIONS was set to ON during the installation, all the SBL programs have been compiled and copied in the target bin directory. If no target was specified, the standard path is assumed (e.g /usr/bin) such that the SBL programs are automatically added to your path. If not, you can add the target bin directory to your path (i.e to add the path to your bin directory to the environment variable PATH).
In the SBL, recall that an application is a package for end-users, providing programs solving a specific biophysical problem, using inputs of different kinds, yet carrying the same semantic.
Program names in the SBL consists in three parts:
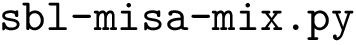
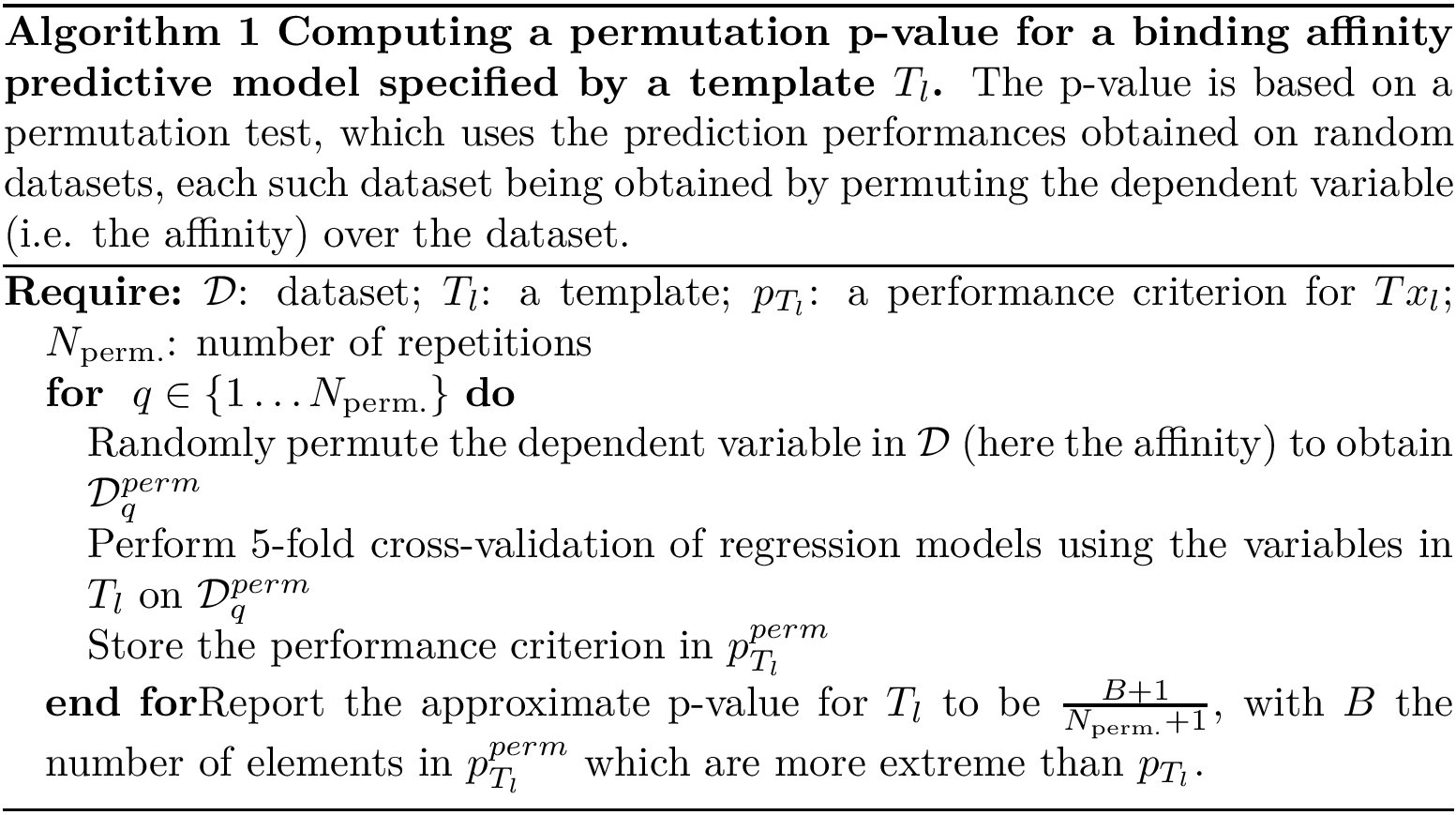
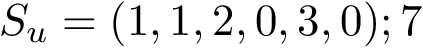
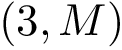
All the programs use a workflow which consists of interconnected Modules. These modules provide command-line options.
The first and possibly most important option is –help that prints on the standard output the list of all options of the program. All the options are grouped by module, with two exceptions:
We now describe the general options common to all programs:
option-name-1 = option_value_1 option-name-2 = option_value_2 ...
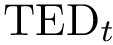
> dot -Tjpg sbl-vorlume-pdb__workflow.dot -o example_workflow.jpg
PDB format. Quoting the PDB, « the PDB format provides a standard representation for macromolecular structure data derived from X-ray diffraction and NMR studies » .
See full details at Protein Data Bank Contents Guide , or an executive summary to deal with coordinates.
One difficulty with PDB files is the lack of topological information, that is chemical bonds between the atoms whose coordinates are listed. For standard amino acids in proteins, the connectivity is naturally known from the chemical structure of each side chain. Non standard connectivity information is provided in the CONNECT section of the PDB file.
HEADER DNA BINDING PROTEIN/DNA 30-APR-01 1IJW
Macromolecular Crystallographic Information File (mmCIF) format. Quoting mmCIF Wikipedia page, the « mmCIF was designed to address limitations of the PDB format in terms of capacity and flexibility, especially with the increasing size and complexity of macromolecular structures being determined. »
See also Beginner’s Guide to PDB Structures and the PDBx/mmCIF Format.
The SBL uses libcifpp to parse PDB and mmCIF files.
There is a vast array of molecular formats, and a reference conversion tool is Open babel.
The focus of the SBL being on biomolecules rather than chemicals, the MOL format is used despite a number of shortcomings.
A precise specification of MOL can be found here.
Creating molecules with the pymol builder. A very convenient strategy to create molecules consists in using the builder from Pymol.
Creating molecules using JSME and Jmol. Another simple strategy to create 3D molecular models of simple molecules operates in two steps. (One can for example follow the following tutorial.)
First, the JSME editor can be used to design the topology of the molecule. A 3D model can then be exported in MOL file format (mouse right click).
Second, the model can be copy-pasted and then edited into a 3D molecular viewer / editor such as Jmol. Within jmol, hydrogen atoms can be added and the structure minimized, and finally exported into the MOL format.
Note also that the File > Get Mol functionality of jmol allows one to pull molecules, as retrieved by the PubChem structure search engine or as resolved by the NCI/NIH chemical structure resolver. As an example, one may download aspirine or caffeine !
Most of the programs from the Applications produce XML files for standard software. This section shows how to process these files:
The XML output files of the SBL programs can be parsed easily using the package PALSE, which provides python functions to load XML files having the same hierarchy of tags, collect selected data with the possibility to use some graphical and/or statistical tools (2D plots, histograms, etc...). Using PALSE reduces a possibly long python script for analyzing the results to few lines.
For a detailed description of the package and a presentation of the main functionalities, the reader is referred to the user manual of PALSE.
The output XML files of the SBL programs may have complicated hierarchies, and / or large numbers of tags. For example, the program 
PALSE offers a simple utility for summing up the common hierarchy of a dataset of XML files. More precisely, given a dataset of XML files, PALSE will load all the XML files, compute for each XML file a tree representing its hierarchy, and compute the largest common subtree of all constructed trees. Two methods allow to access to this common hierarchy in two different ways:
Hence, visualizing the common hierarchy is simply done by printing the output of one of these methods.
Once the hierarchy is obtained, one wants to analyze the dataset of XML files: PALSE offers a simple methodology in three steps:
Most of the programs from the Applications produce visualization files for standard software. In the following, we comment on various visualization facilities:
The visualization softwares are used to visualize various output results, be they collections of atoms extracted from PDB files, or geometric constructions related to the particular problem being solved (molecular interfaces, binding patches, coarse-grain approximations, etc). When a calculation generates an output to be visualized, an option containing the keyword viewer is available. Such an option can be set to none (no output), vmd (VMD) or pymol (PyMOL). Since visualization files may be heavy, those options are set to none by default. If set for one of the aforementioned systems, a visualization file of the corresponding format will be produced when reporting the results.
When visualizing the results using a visualization software, two kind of objects fundamentally different may be seen:
While the selections are dynamical (once it is loaded, a user can modify the selection), the geometric objects are static (once it is loaded, the geometric objects cannot be modified).
VMD is a molecular visualization program for displaying molecules using 3-D graphics. In addition to the visualization files in VMD format, the SBL library also provides a number of plugins for VMD that can be automatically installed as specified in section Installing VMD plugins . Two important plugins are:
The program PyMOL is a molecular visualization system on an open-source foundation. See PyMOL user manuals for more details. The visualization files for PyMOL are python scripts that can be run directly from PyMOL .
Graphviz is a graph visualization software especially useful to plot various types of diagrams . Input files follow the dot file format, and different programs allow to generate pictures of the graphs such as dot (for a hierarchical embedding), circo (for a circular embedding), etc... The dot file format is used in our Applications when an output is a graph, or for visualizing the workflow of an application.