 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
Packages | |
| Alphafold_analysis | |
 | Here you should put an abstract of your package Reference Manual – User Manual . Authors: F. Cazals and E. Sarti |
| Binding_affinity_prediction | |
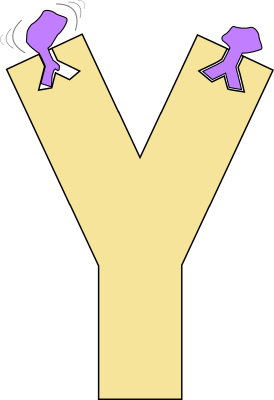 | Binding_affinity_prediction is package implementing a complete workflow to predict affinities from geometrical and biophysical parameters using bound and unbound crystallographic structures. It generates and evaluates linear models using various combinations of these parameters and selects the most accurate ones. Reference Manual – User Manual . Authors: S. Marillet and P. Boudinot and F. Cazals |
| FunChaT | |
 | A package to build biased HMMs by exploiting the results of Structural motifs. Reference Manual – User Manual . Authors: R. Tetley and F. Cazals |
| Genetrank | |
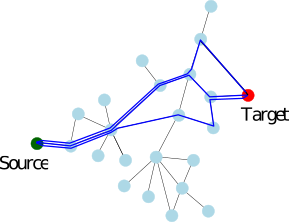 | Gene ranking based on random walk with restarts in Protein Protein Interaction Networks Reference Manual – User Manual . Authors: F. Cazals and A. Jean-Marie and D. Mazauric and J. Roux and G. Sales Santa Cruz |
| Molecular_cradle | |
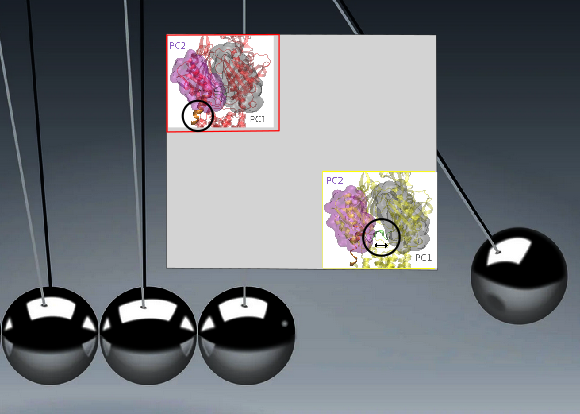 | A package to identify almost rigid regions in complex biomolecules Reference Manual – User Manual . Authors: M. Simsir and F. Cazals |
| Multiple_interface_string_alignment | |
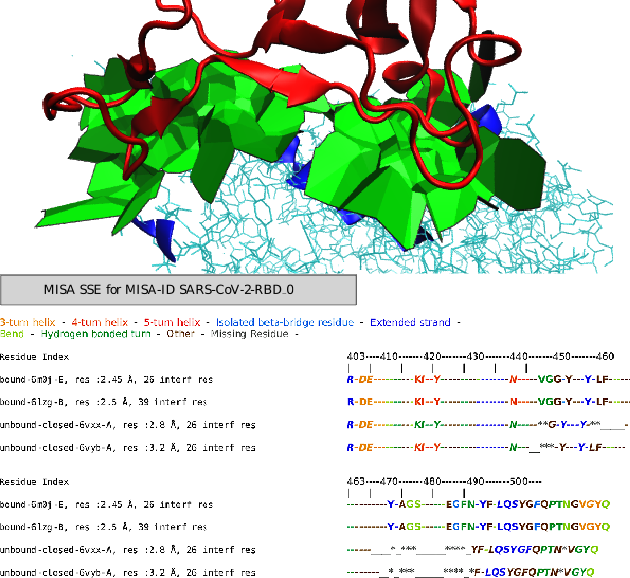 | MISA provide a coloring of interface strings ie amino acids found at interfaces with various biological and biophysical quantities. Reference Manual – User Manual . Authors: S. Bereux and F. Cazals |
| PDB_utilities | |
 | Various utils providing simple manipulations on PDB files Reference Manual – User Manual . Authors: F. Cazals |
Applications combining several ingredients from the previous groups.
None.