 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
Authors: F. Cazals and T. Dreyfus
Rationale. Numerous applications resort to the same data processing. For example, computing the volume of a molecular models merely requires computing the volume of a union of balls, whatever the semantics (atoms, pseudo-atoms) of these balls. To make such routine operations available on the shelf, we use modules. Modules are small pieces of C++ code which can be chained within a workflow to define a whole application. In short, a module is characterized by three main features:
Importance of modules and workflow. Connecting modules using a directed graph which defines a workflow yields immediate key functionalities:
Overview. In the following, we first explain how to customize existing applications using modules and workflows in section Customizing Applications. We then explain how to create one's workflow in section Developing Workflows with Existing Modules, and finally how to create new modules in section Developing Modules.
To go straight to the point, let us review the main steps involved in defining an application, based on modules and a workflow:
Step 1. For each class performing a significant step in the application, one defines the module associated to this class. One can consult SBL: existing modules
Step 2. Because our goal is to handle coherently cases where the objects manipulated are the same – yet with different semantics (see the example of balls in Introduction), we assemble within the workflow traits and the workflow classes the required types
Step 3. Assemble the application by successively
instantiating the modules.
defining the workflow traits using the types of modules.
defining the workflow
See the example below.
Applications in the SBL intensively use modules to re-use core components involved in several applications, and to easily customize a given application to custom data models. Static versions of the executables are available at the SBL Applications page, or can be compiled directly from the library using CMake .
From the end-user standpoint, modules offer handy features, namely the possibility to :
display the workflow of the application, in order to see the main steps (i.e modules) of the algorithm used in the application,
display the help which groups the options by steps of the application.
In addition to specific options inherent to a given application, the following generic options—implemented in the workflow data structure—are offered for all applications:
–help : prints on the standard output all the command-line options of the application, with a brief description.
–config-file : reads the command-line options from a configuration file as specified in the Boost Program Options documentation.
–workflow : prints the workflow as a directed graph, namely .dot file to be visualized with Graphviz. The command for converting the .dot file onto a .pdf file is printed on the standard output.
–verbose : dumps high level statistics:
0 : default option: no output gets printed on the console.
1 (run level): the program prints intermediate statistics on the tasks performed.
2 (final statistics): the program prints the final statistics.
–log : Redirects the standard output into a log file (except for the help and the workflow). It sets the verbose level to final statistics, unless the verbose level is specified separately.
–colored-log colors the log (red for module and loader names, green for statistics, blue for report).
–directory-output : enforces the creation of all output files into the specified directory. NB: the directory must exist.
–output-prefix : if specified with an argument, uses the specified prefix for all output files; if no prefix is specified, generates an output prefix by concatenating the specified option names and the associated values.
–uid : adds to the end of the output prefix a unique identifier, corresponding to the local date and time, at the micro-second time scale.
–report-at-end : reports all the output files after the execution of all modules in the workflow, rather than after each module execution.
The two options –output-prefix and –uid add a common prefix to all output files of a given execution, identifying the output of a process. The case study is as follows:
| Without uid | With uid | |
| Without output prefix | <application-name> | <application-name>__<uid> |
| With implicit output prefix | <application-name>__<input-option-values> | <application-name>__<input-option-values>__<uid> |
| With explicit output prefix | <explicit-prefix> | <explicit-prefix>__<uid> |
In C++, a traits class is a data structure defining types. In generic programming paradigm, traits are used to group the definitions of types used in template classes.
In our case, template classes are the modules of the SBL, and the traits classes used to instantiate them provide the required types.
All applications in the SBL were developed following the same guidelines :
Workflow class. The workflow base class SBL::Modules::T_Module_based_workflow specifies the common features to all workflows (building the workflow, executing the modules within the workflow, adding common command-line options to a program, etc.). This workflow template class is parameterized by a traits class, and the workflow class of any application inherits from it.
Traits class. A Workflow class uses a single traits class defining all the types used by the modules in the workflow. The type definitions are ascribed to two categories : the biophysical models and the other types; generally, all biophysical models are conceptualized, i.e they are the template parameters of the traits class–that is the traits class is itself a template class.
Thus, adapting an existing application to a given biophysical model merely requires modifying the biophysical model definition in the source. To ease this process, these biophysical models generally match biophysical concepts in Models, so that the corresponding concept has a full documentation (user manual and reference manual), and a list of predefined models.
As an example of workflow, consider the application Space_filling_model_interface_finder, which computes the binary interfaces between the chains or domains of a molecule, and outputs a graph representing those interfaces. The workflow for this application is displayed just below. The traits class of the workflow is T_Space_filling_model_interface_finder_traits and is templated by four concepts:
ParticleTraitsBase : class providing a base representation of a particle (atom or pseudo-atom), see ParticleTraits ;
PartnerLabelTraits : class providing a hierarchy of labels to assign the particles to partners. Interfaces are indeed sought between partners identified by these labels. See details in MolecularSystemLabelsTraits ;
ParticleAnnotator : class providing an annotation system attached to the particles, either for annotations directly used in the calculations (e.g the radii of particles), or for user defined annotations not used for calculations but delivered in the output files, see ParticleAnnotator ;
ParticlesBuilder : a functor (i.e a class providing the operator "()") building the particles from an input data structure to the data structure provided by ParticleTraitsBase , see ParticleTraits .
Each of these concepts have models provided by packages in Models, so that producing programs of the same application in a different context could be a matter of replacing one line in the corresponding source file.
We now discuss how to create an application using modules, which encompasses 3 main steps:
In the sequel, we provide a simple program using modules, and further discuss the three steps just described.
This first example involves a toy program using a workflow with only one module.
The program just creates a grid 100x100 of 2D points, and store the points in a spatial search engine–to be used for example to report the nearest neighbors of a given point. It uses in particular the module from the package Spatial_search. This module is templated by a Traits class defining two types :
Points_container : the type representing the container of 2d points,
To embed a module into a workflow, we define a class inheriting from SBL::Modules::Module_based_workflow, which
implements a directed graph connecting modules while taking care of numerous features,
The following example illustrates these steps on our simple application:
Executing this program without any option yields the help of the program. An effective execution requires at least one option to be specified, e.g the –verbose option (which is off by default).
Of particular interest is the –workflow option, which does not execute the workflow but instead creates a Graphviz file representing the workflow. Once processed by Graphviz, in image file of the workflow is obtained, allowing to an overview of the application at a glance.
In the sequel, we detail the aforementioned steps.
The workflow's traits class is described in section Customizing applications: workflow's traits class .
Registering a module consists in creating the corresponding node in the workflow graph. This is done using the method SBL::Modules::Module_based_workflow::register_module .
This step consists in specifying which module(s) are executed first. This is done using the method SBL::Modules::Module_based_workflow::set_start_module. Note that the workflow can start with multiple modules rather than one : in such a case, the execution order of these modules is arbitrary.
In the example of section A simple example , the attributes of the module have been initialized by directly setting them. It actually turns out that there are three ways to initialize the input of a module:
Initialization by setting attributes. One just calls a function modifying attribute(s) of the module.
Initialization via the connection of two modules. In this case, the output of a source module becomes the input of a target module.
Initialization from loaded data. In this case, the initialization involves data loaded from a file into the module, the required information (filename, possible options) coming from the command-line options.
We now detail the latter two options.
Initialization via the connection of two modules.
This is done using the method SBL::Modules::Module_based_workflow::make_module_flow . This method takes four arguments : the vertex representing the source module, the vertex representing the target module, a linker function allowing to initialize the target module from the source module, and optionally a name to display over the arc representing the connection in the workflow's graph display.
Initialization from loaded data.
Data can be loaded from files using special classes called loaders. The framework of modules provides a number of loader classes loading data from a file into main memory. All loaders specialize the virtual class SBL::IO::Loader_base by possibly re-implementing the method SBL::IO::Loader_base::load . A loader can be independently used, or can be stored as an external property of the workflow graph. In doing so, command-line options related to the data to be loaded are added to all other command-line options of the workflow. Storing a loader as an external property is done using the method SBL::Modules::Module_based_workflow::add_loader
The following program loads the points from a file specified in the command-line options, and fills the search engine :
Two things should be noticed :
calling the method SBL::Modules::Module_based_workflow::load results in the execution of the load method of each loader. Note that the call order of the loaders is arbitrary.
the new method initialize sets the loaded points from the loader to the spatial search module right after the call of the method SBL::Modules::Module_based_workflow::load
Note that the helper of the executable has been enriched with the options provided by the loader.
This section describes the algorithm processing the workflow, the semantics of the vertices and edges of the workflow, and how to alter the default behavior of the workflow processing.
The workflow is represented by a directed graph where vertices are modules and arcs are connections between modules. Note that the graph contains also the start vertex that is not associated to any module : it is used just as a start point for the workflow processing. As mentioned in section Introduction , the algorithm used for processing the workflow is a variant of a depth first traversal algorithm. The algorithm uses a recursion stack initialized with the start vertex, and executes the following steps while the stack is not empty :
As we shall see below, this basic algorithm can be tuned using specific modules and the associated operations. To describe them, we first list the data structures used for vertices and edges of the workflow.
what are the different vertices (module_base, predefined, user defined, etc...)
same for edges
Operations are functionality of the workflow that alter the base behavior of the traversal algorithm. These operations are necessary for fully representing the SBL applications. There are five operations, represented by keywords :
Except the OR operation, all operations modify the algorithm behavior. In the following, we describe how the algorithm is modified for each operation, and how to use these operations.
Rationale.
Remind that when a module have several in-going modules, it is visited immediately after execution of one of its in-going module, corresponding to the OR operation. However, steps of the workflow may require that all steps before have been already executed. To enforce the visit of a module only after all its in-going modules have been visited, one has to resort on conjunction modules.
Use case.
A conjunction module is represented by the class SBL::Modules::T_Module_conjunction . Practically, the AND operation requires
SBL::Modules::Module_based_workflow::Vertex conj = this->make_conjunction(u, v);
Accessing the in-going modules is done through the methods SBL::Modules::T_Module_conjunction::get_conjunction_1 and SBL::Modules::T_Module_conjunction::get_conjunction_2 .
Rationale.
In a workflow, some of the steps may be optional, i.e some modules should be executed only if a user specifies so. We need a way to add a command-line option for the user to specify if the module should be executed or not.
Use case.
A boolean tag is associated to all vertices of the workflow's graph via an external property. This association is done using a map from the vertices to the tag. Note that vertices are represented by successive integers so that access to the tag of a vertex is done in constant time. When processing the stack of the workflow, if the tag of the top vertex is false, the vertex is simply discarded with no processing.
By default, the tag of all vertices is true. However, when a module is optional, the default value of the tag is false. It is only after the command-line options are parsed that a tag of an optional module can be set to true.
An existing module is made optional using the method SBL::Modules::Module_based_workflow::make_optional_module . This method turns the default tag value to false, and adds a corresponding command-line option. Considering the previous example on spatial search engines, to make the module Spatial_search_module optional, the following line is added to the construction of the workflow in the previous example :
this->make_optional_module(v, "search-engine", "Run the spatial search module.");
Rationale.
To implement an If-Then-Else on modules, we use a specific module equipped with a predicate, so as to choose which next module should be executed.
Use case.
A conditional module is created with the method SBL::Modules::Module_based_workflow::make_condition
This previous method creates an object of type SBL::Modules::T_Module_condition , which has up to two out-going modules (to implement an If or an If-Then-Else).
The method SBL::Modules::Module_based_workflow::make_condition requires up to height arguments :
the vertex u representing the in-going module in the workflow,
the predicate P that is a functor taking as argument a pointer to to the in-going module, and returning a boolean value;
the vertex v corresponding to the action to be executed if the predicate evaluates to True,
the updater of the input from u to v (as the functor in the method SBL::Modules::Module_based_workflow::make_module_flow),
(optional) the vertex w corresponding to the action to be executed if the predicate evaluates to False,
(optional) the updater of the input from u to w (as the functor in the method SBL::Modules::Module_based_workflow::make_module_flow),
(optional) a name for the predicate for displaying the condition module in the printed workflow, if any,
The following example involves a conditional loop to build an approximate spatial search engine for a collection of 2D points (loaded from a file here), meeting specific criteria. It implements a module computing the sum, over all points from the DB, of the distances each point to its nearest neighbor. If the sum is less than a threshold, the algorithm halts; otherwise, it rebuilds the search engine.
Two used features call for the following comments:
The class Is_invalid_engine is the predicate governing the re-construction: its input is the module before the condition, and it uses the output of this module for computing the predicate value.
The method SBL::Modules::Module_based_workflow::make_condition creates the conditional module and the arcs linking the different involved vertices. Note that there is no module associated to the Else statement – nothing happens if the predicate evaluates to False. Note also that the method has explicit template parameters which are the input module type, and the output module type. In the case where a second output module exists, its type is added to the template parameters of the method.
Rationale.
We wish to repeat the execution of a given module on a collection of input data. This objective is achieved by creating a collection of instances of the module of interest.
Use case.
A collection of modules is an instance of the template class SBL::Modules::T_Modules_collection < Module , SetIndividualInput > . Such a module is registered with the method SBL::Modules::Module_based_workflow::register_module . The template parameters are as follows:
When the collection of modules is initialized, the method SBL::Modules::T_Modules_collection::set_individual_inputs calls the previous functor for each of its input data (using an iterator). For each such input, a new instance of the module of interest is created (dynamic instance created in C++ with new), and initialized with this input using the functor SetIndividualInput . Note that the type of the second parameter of the functor must match the value type of the iterator.
When the collection of modules is executed in the workflow, each instance of the modules in the collection is executed.
One can iterate on the created modules with the methods SBL::Modules::T_Modules_collection::modules_begin and SBL::Modules::T_Modules_collection::modules_end, or access them directly with the method SBL::Modules::T_Modules_collection::get_module .
The following example loads a set of 2D point collections, and builds a spatial search engine for each such collection.
Two used features should be stressed:
the class Set_points is the initialization functor. Note that it always has two arguments, the first one being the module, the second one being a user-defined data structure containing the input.
In developing a module, several virtual functions can be customized by developers.
Implementing a module requires specializing the base class SBL::Modules::Module_base, which implements all basic functionalities. The class SBL::Modules::Module_base is a pure virtual containing the pure virtual method SBL::Modules::Module_base::run. This method is the one called to execute the module within the workflow, and thus needs to be implemented.
In the sequel, we present the remaining functionalities of modules. Note that these functionalities are virtual methods from the class Module_base. Thus, as virtual methods, they may be redefined by the programmer.
SBL::Modules::Module_base::statistics : printing the statistics of the run.
After a module has been executed, it is possible to group all the statistics related to the calculations in the method SBL::Modules::Module_base::statistics . In particular, this method is called only for particular values of the verbose mode (2 and 3). This is particularly useful for time-consuming statistics. By default, this method does nothing.
SBL::Modules::Module_base::is_runnable : checking if a module can be executed.
When particular cases are not handled, the execution of a module may not produce the expected output : in such a case, the modules downstream cannot be executed. The method SBL::Modules::Module_base::is_runnable provides a mechanism to check whether the input of a module is correctly set. By default, this method returns always true.
SBL::Modules::Module_base::report : reporting module output into files.
Recording output data of a module into files is done re-implementing the method SBL::Modules::Module_base::report. The argument of this method is the prefix generated by the workflow class (see section Customizing Applications). By default, this method does nothing.
Note that the method SBL::Modules::Module_base::report is not called by default in the workflow : to be called, the module has to be set as an "end" module, meaning that it produces final output that should be recorded. To set a module as an "end" module, the workflow method SBL::Modules::T_Module_based_workflow::set_end_module should be used on the vertex representing the module. It is also possible to change the way the workflow reports the output into files through the command-line option –report-at-end: either right after the execution of a module (default), either after all modules have been executed.
SBL::Modules::Module_base::set_module_instance_name : naming duplicated modules.
When duplicating a module in the workflow (e.g, within a collection of modules), the output prefix provided by the workflow for reporting the output of a module is the same for every modules of the collection : this means that the reported data will be overwritten while reporting the different instances of the same module.
In that case, a specific name can be given to each instance using the method SBL::Modules::Module_base::set_module_instance_name . A given instance can then be retrieved using the method SBL::Modules::Module_base::get_module_instance_name . This is also useful for identifying the instances on the log during the execution of the workflow. By default, this method returns the empty string.
SBL::Modules::Module_base::get_name : interacting with modules documentation in the Applications user manuals.
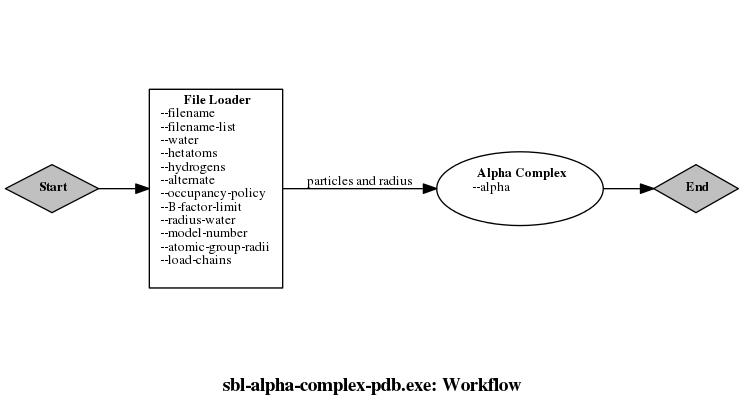
Each user manual from Applications generally provides a workflow interactive image of the concerned application. This image is based on Graphviz and corresponds to a ".dot" file produced by the executables of the application itself (see command-line option –workflow).
By interactive, we refer to the fact that the image contains hyper-references for each node (i.e. module) of the workflow. By default, every hyper-link points to the user manual of Module_base. To specify another hyper-reference, e.g the reference manual or the user manual of the module, one needs to redefine the virtual method SBL::Modules::Module_base::get_name, returning the string corresponding to the class name or to the package name. The corresponding manual is inferred from this string.