 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
Authors: F. Cazals and T. Dreyfus
In the SBL, a number of low-level algorithms are based upon combinatorial, geometric and topological representations, so that these algorithms do not require any specific biophysical information. This fact allows developing several programs taking as input the same geometric objects, yet, in different biological contexts.
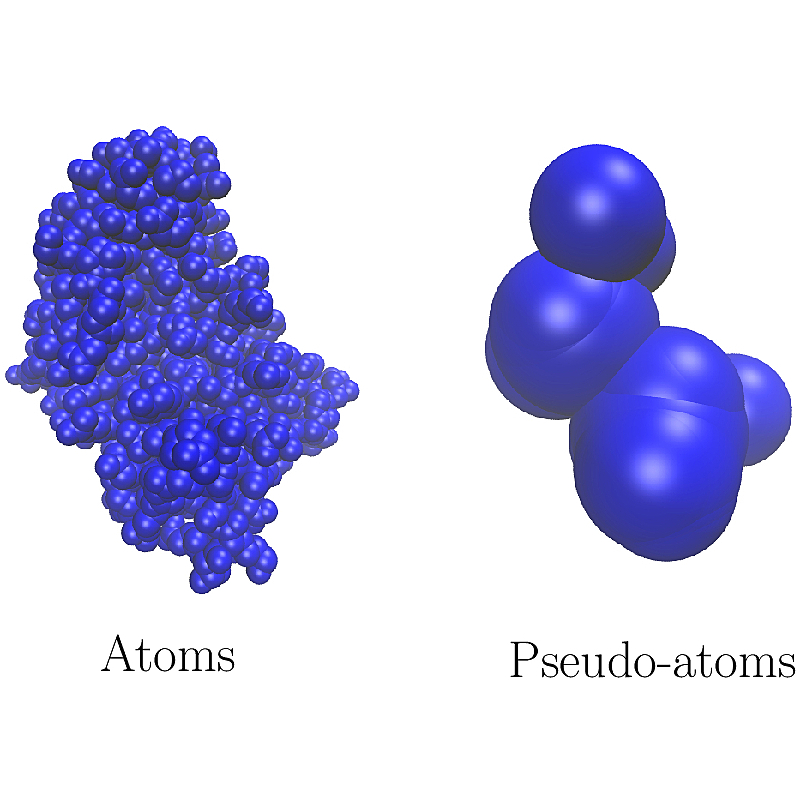
The iconic example is the calculation of molecules surfaces and volumes: such a calculation requires a set of balls, which may be atoms or pseudo-atoms. For this reason, the SBL proposes two programs, namely 
In fact, the very generic nature of prerequisites on objects makes it possible to use very generic algorithms within the SBL.
A particle is an elementary physical object. It is of geometric nature, but is potentially endowed with other properties. To handle these pieces of information, the ParticleTraits package define classes to:
From the user standpoint, using programs resorting to models of ParticleTraits has two implications:
To ease things, when an application is instantiated with several models of ParticleTraits, the name of the corresponding programs contains a key-word identifying the type of the particle (see section Example: Running a Program using Particles).
The following example shows the differences between two programs of an application using two different models of the concept ParticleTraits. Let consider the programs of the application Space_filling_model_surface_volume, that computes the volume and the surface area of an input molecule. The input molecule may be stored in files with different formats, as the PDB file format, or as a simple list of 3D spheres in a plain text file. Depending on the format, the way to load and to represent a particle will not be the same, so that two programs are provided:

sbl-vorlume-pdb.exe -v -c -f demos/data/1vfb.pdb
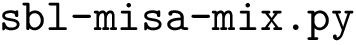
sbl-vorlume-txt.exe -v -c -f demos/data/spheres.txt
The output files of 
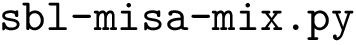
The class SBL::Models::T_Geometric_particle_traits< GeometricKernel , GeometricRepresentation > defines a type of particle that is purely geometric and has no attached biophysical information.
The geometric part of the SBL is based upon the CGAL library: the template parameter GeometricKernel determines the geometric primitives used for representing a particle (number type, point type, etc...), as explained in the documentation of the CGAL library. The second template parameter GeometricRepresentation is the geometric representation of the particle, which may be represented by CGAL objects such as Weighted_point, Point_3, Sphere_3. The default geometric representation used is the Weighted_point type from CGAL type.
The SBL provides two main data structures for handling atoms, that are presented just below:
The ESBTL provides the hierarchical data structure ESBTL::Molecular_atom to represent an atom in a hierarchical manner–namely the atom belongs a residue which is itself in a chain which itself belongs to a molecular model. The SBL provides the model SBL::Models::T_Atom_with_hierarchical_info_traits< GeometricKernel , SystemItems > for using the class ESBTL::Molecular_atom within the SBL algorithms. The template parameter GeometricKernel is a class defining the number type FT used to represent the coordinates of the atoms, and the type Point_3 used to represent the atoms themselves as geometric entities. It has a default type for defining geometric objects with float coordinates and calculations. The template parameter SystemItems is a class defining the base classes used to represent atoms, residues, chains and models in a molecule (see the ESBTL documentation). It has the default type from the ESBTL library.
Due to the previous remark, in order to serialize an atom, the entire molecule containing it has to be serialized. For this reason, in the SBL, the ESBTL Molecular Atom can only be partially saved in a file, and cannot be loaded from a file. In order to have a completely serializable data structure, the SBL provides a "flat" data structure, in the sense that all the information on the atom is contained in the atom. The SBL provides the model SBL::Models::T_Atom_with_flat_info_traits< GeometricKernel , SystemItems >. The two template parameters are the same ones as explained in section ESBTL Molecular Atom as Particle Type .
The ESBTL provides a pseudo-atom data structure ESBTL::Coarse_atom to represent residues with a fix number of pseudo-atoms. As for the ESBTL atom data structure, the ESBTL pseudo-atom data structure is not fully savable: the SBL provides a "flat" pseudo-atom data structure SBL::Models::T_Pseudo_atom_per_residue_spec_with_flat_info and a model of ParticleTraits for this pseudo-atom class: SBL::Models::T_Pseudo_atom_per_residue_spec_with_flat_info_traits< GeometricKernel , SystemItems >.
As explained in the user manual of the package ParticleAnnotator, a particle may be decorated with annotations: these annotations may be compulsory or optional, depending on the context.
The class SBL::Models::T_Particle_with_annotations_traits< ParticleTraitsBase , AnnotationsType > inherits from the base traits class ParticleTraitsBase and re-implements the class Particle_type by simply adding an annotations attribute of type AnnotationsType .
Each of the predefined particle traits class without annotation has its own version with annotations, using the class SBL::Models::T_Particle_with_annotations_traits:
In any case, such classes have fixed annotations inheriting from the template parameter Annotations (if not void) such that:
Note that all the corresponding traits classes define also a Default_annotator type that is the annotator type for the default annotations (the template parameter Annotations is void).
As explained in the user manual of the package MolecularSystemLabelsTraits, when particles are part of a molecular structure, the particles are associated to one of the partner of the structure, or to interfacial water, or even to extra particles. The class SBL::Models::T_Particle_with_system_label_traits< ParticleTraitsBase , PartnerLabelsTraits , MediatorLabelsTraits , ExtraLabelsTraits , IsSerializedLabel > provides a type of particle enriched with a system's label. The template parameters are, in their order of appearance:
This example shows the source file of the program 
This source file is organized as follows:
Note that it is sufficient to change the models used in (i) for making a new program with a new type of particle. When particles are atoms, one may use the model SBL::Models::T_Atom_with_flat_info_and_annotations_traits as shown:
When particles are simple 3D spheres, one may use the model SBL::Models::T_Geometric_particle_traits as shown:
The C++ concept ParticleTraits has five requirements:
This example shows the model SBL::Models::T_Geometric_particle_traits of the concept ParticleTraits representing a particle as a simple 3D geometric object (a weighted point by default):
While the three main types discussed in section Developing new models of ParticleTraits concept are defined in this header file, the serialization of the default type (the weighted point) is included from another header file (SBL/IO/Weighted_point_serialize.hpp), and the weighted-points are by default drawable using the package Molecular_viewers.
There is an additional type SBL::Models::T_Geometric_particle_traits::Annotator_default that is not a requirement but that is used for simplifying the code when instantiating an application with several models of ParticleTraits.