 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
Authors: F. Cazals and T.O'Donnell
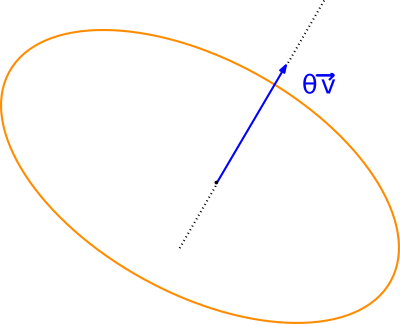
There are several ways to define and parameterize 3D rotations, see e.g. Wiki/rotation in 3D and Wiki/rotation matrix :

This package provides various parameterization of rotations using these formalisms, using low level classes from Eigen.
The reader is referred to [8] as well as rotations 3D formalism and rotation matrix .
We also note in passing that there exist various metrics between 3D rotation, see [126] and [109] .
3D Rotations are performed on points trough a single overloaded functor templated by a number type: SBL::GT::T_Rotation_3
This class takes in as input from one to three angular radii values followed by the same number of orthogonal axes around which the corresponding rotations are applied. If no axes are given the unity vectors x y and z are used by default(in this order). Varying representations of 3 dimensional rotation can be obtained through methods of SBL::GT::T_Rotation_3:
The functor operator needs an iterator input for points and an iterative output to apply the rotation directly. Finally if on top of all the previous parameters a center of rotation is supplied then the points will be centered before rotation is applied.
The following examples shows how to rotate a set of randomly drawn 3D points: