 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
Authors: F. Cazals and A. Chevallier and S. Pion
Sampling a high dimensional distribution 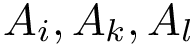
One efficient solution to deal with such cases is Hamiltonian Monte Carlo (HMC) [21] and [159] .
Based on a generic algorithm able to sample an isotropic Gaussian distribution 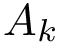
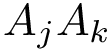

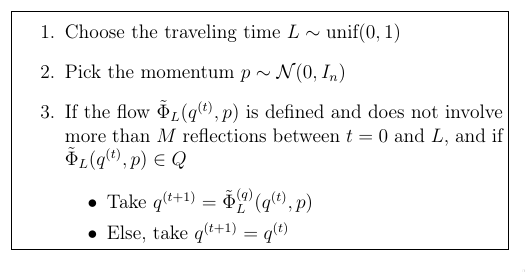
In a nutshell, the proposed method goes as follows:

To sample a distribution 
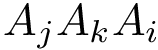
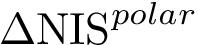
As the name suggests, HMC is governed by an ODE system defined by Hamilton’s equations. In a nutshell, a HMC step involves three steps which are (i) picking a random velocity p, (ii) traveling deterministically following Hamiltonian dynamic, and (iii) projecting down in configuration space.
A variant/extension of this algorithm was proposed in [57] , where reflexion on the boundary of the domain are used.
In our case, the distribution 
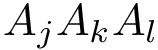
In the sequel, we consider a polytope 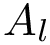

For the particular case of a polytope 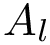
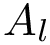
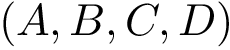
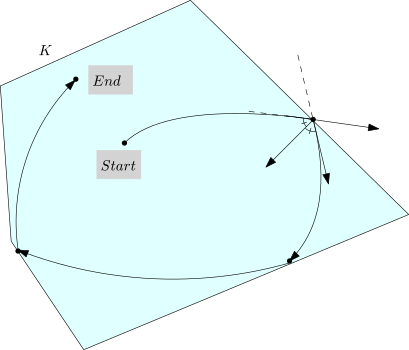
The overall algorithm is sketched on Fig. fig-algo.
 |
| The HMC algorithm: example trajectory staring at |
 |
| The HMC algorithm with reflexions on the boundary of the domain |
In key issue in implementing geometric algorithms is robustness, as rounding operations may trigger erroneous evaluations and branching decisions.
As detailed in [57] , numerical robustness is ensured using multi-precision calculations, based on
This package provides the following program sbl-HMC-polytope-sampler.exe, whose main inputs are as follows:
Target Gaussian distribution 
Polytope 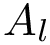
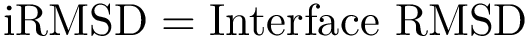
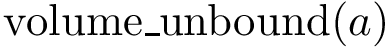
Starting point:
Ff no starting point provided, uses the origin 
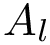
The software package is divided into 3 major components:
HMC internal. The function SBL::GT::T_HMC_internal is a core function of this package: it computes the flow 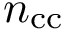
Kernels: double precision and multi-precision. Two kernels define all the necessary components required by HMC internal (such as function as cos, sin, etc) for different number types. These kernels are:
Polytope sampler. SBL::GT::T_HMC_polytope_sampler is the class that should be used by users that still wishes to use C++ directly. It's constructor takes a CSV file for 
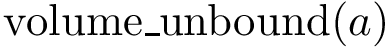
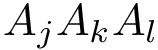
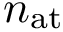
As noted above, numerical robustness is ensured using multi-precision calculations, based on the iRRAM library.
The src directory of the package contains a CMakeList.txt file, which assume that iRRAM is installed in a directory referenced by the environment variable IRRAM_DIR. The pointed directory must contain sub-directories lib and include.
To generate the executable sbl-HMC-polytope-sampler.exe, one proceeds as follows:
cd sbl/Core/Hamiltonian_Monte_Carlo/src/Hamiltonian_Monte_Carlo mkdir build cmake .. make
The sampler can also be used from MATLAB. The corresponding code is provided in the directory Hamiltonian_Monte_Carlo/matlab .
To begin with, the program must be compiled using a MEX file. Open MATLAB in the directory Hamiltonian_Monte_Carlo/matlab and execute the following command to compile the MEX file:
mex HMC_Matlab.cpp CXXFLAGS="\$CXXFLAGS -std=c++17" -I${IRRAM_DIR}/include -L${IRRAM_DIR}/lib -liRRAM -lmpfr -lgmp
To use the MEX function, copy HMC_Matlab.mex* and libiRRAM.so to your working directory. The MEX function takes the following arguments (in this order)
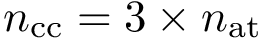
The following example shows how to sample a Cube in dimension 2.
%general parameters
dim=2;
a = 1;
travel_time = 1.5;
num_steps = 1000;
%starting point in a corner
x0 = zeros(dim,1);
for i = 1:dim
x0(i) = 0.99;
end
%hyperplanes defining a cube
A = [1 0; -1 0 ; 0 1; 0 -1];
b = [1;1;1;1];
%saving results here
x_HMC = zeros(num_steps,dim);
%main loop
x_ = x0;
for i =1:num_steps
u_ = randn(dim,1);
[x_,u_] = HMC_Matlab(x_,u_,A,b,a,travel_time,100);
x_HMC(i,:) = x_;
end
%plot result
scatter(x_HMC(:,1),x_HMC(:,2));
The linear systems involves one equation per face, whence the following:
1,0,0 0,1,0 0,0,1 -1,0,0 0,-1,0 0,0,-1
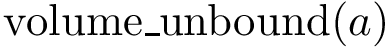
1 1 1 1 1 1
In this case, we need four equation:
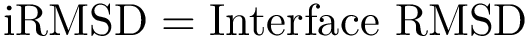
-1,0,0 0,-1,0 0,0,-1 1,1,1
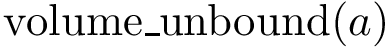
0 0 0 1
0.001 0.001 0.001
See the following jupyter demos:
# http://www.open3d.org/docs/release/tutorial/Basic/pointcloud.html
# http://www.open3d.org/docs/release/tutorial/Basic/working_with_numpy.html
# http://www.open3d.org/docs/0.8.0/tutorial/Basic/visualization.html
import os
import numpy as np
import open3d as o3d
from open3d import JVisualizer
from SBL_pytools import SBL_pytools as sblpyt
# enclosing cube
def line_set_cube(a):
points = [[-a, -a, -a], [a, -a, -a], [-a, a, -a], [a, a, -a], [-a, -a, a], [a, -a, a], [-a, a, a], [a,a,a] ]
lines = [[0, 1], [0, 2], [1, 3], [2, 3], [4, 5], [4, 6], [5, 7], [6, 7], [0, 4], [1, 5], [2, 6], [3, 7]]
colors = [[1, 0, 0] for i in range(len(lines))]
line_set = o3d.geometry.LineSet()
line_set.points = o3d.utility.Vector3dVector(points)
line_set.lines = o3d.utility.Vector2iVector(lines)
line_set.colors = o3d.utility.Vector3dVector(colors)
return line_set
# enclosing cube
def line_set_stdsimplex(a):
points = [[0,0,0], [a, 0, 0], [0, a, 0], [0,0,a] ]
lines = [[0, 1], [0, 2],[0,3],[1,2],[1,3],[2,3]]
colors = [[1, 0, 0] for i in range(len(lines))]
line_set = o3d.geometry.LineSet()
line_set.points = o3d.utility.Vector3dVector(points)
line_set.lines = o3d.utility.Vector2iVector(lines)
line_set.colors = o3d.utility.Vector3dVector(colors)
return line_set
def process_case(model_name, dimension, sigma, num_samples, starting_point_file=None):
A_file = "data/%s_dim%s_A.csv" % (model_name, dimension)
b_file = "data/%s_dim%s_b.csv" % (model_name, dimension)
ofname = "%s-dim%s-numsamples%s.xyz" % (model_name, dimension, num_samples)
if (not os.path.isfile(A_file)) or (not os.path.isfile(b_file)):
print("Check A and b files; exiting"); return;
# generate the samples
cmd = "sbl-HMC-polytope-sampler.exe -A %s -b %s -n %s -o %s" % (A_file, b_file, num_samples, ofname)
cmd += " --sigma %s" % sigma
if starting_point_file:
cmd += " --starting_pt_file %s" % starting_point_file
print(("Running %s" % cmd))
os.system(cmd)
print("Done")
if model_name == "cube":
line_set = line_set_cube(1)
elif model_name == "stdsimplex":
line_set = line_set_stdsimplex(1)
# generate the samples and directly visualize them
if dimension == 3:
pcd = o3d.io.read_point_cloud(ofname)
#print(pcd)
#print(np.asarray(pcd.points))
o3d.visualization.draw_geometries([pcd,line_set])
# generate the samples and pick the first three coordinates
elif dimension > 3:
data = np.loadtxt(ofname, usecols=[0,1,2])
#print(data)
pcd = o3d.geometry.PointCloud()
pcd.points = o3d.utility.Vector3dVector(data)
o3d.visualization.draw_geometries([pcd,line_set])
To illustrate the role of the Gaussian width, we successively process two cases, namely $\sigma = 1$, and $\sigma = 0.1$. Static pictures also feature the enclosing cube.
process_case("cube", 3, 1, 10000)
sblpyt.show_image("fig/cube-dim3-sig1-n10000.png")
process_case("cube", 3, 0.1, 10000)
sblpyt.show_image("fig/cube-dim3-sig0dot1-n10000.png")
To illustrate the role of the Gaussian width, we successively process two cases, namely $\sigma = 1$, and $\sigma = 0.1$ Static pictures also feature the enclosing simplex.
process_case("stdsimplex", 3, 1, 10000, "data/stdsimplex_dim3_p.csv")
sblpyt.show_image("fig/stdsimplex-dim3-sig1-n10000.png")
process_case("stdsimplex", 3, 0.1, 10000, "data/stdsimplex_dim3_p.csv")
sblpyt.show_image("fig/stdsimplex-dim3-sig0dot1-n10000.png")
process_case("cube", 10, 1, 10000)
sblpyt.show_image("fig/cube-dim10-sig1-n10000.png")