 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
Authors: F. Cazals and T. O'Donnell
Centers of mass and their generalization.
The celebrated center of mass of a point set in a Euclidean space is the (a) point minimizing the sum of squared Euclidean to points in this set.
Similarly, the 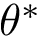
This package provides a method to compute these quantities for points located on the unit circle 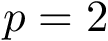
Definitions. Consider 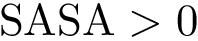
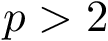
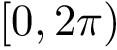
The geodesic distance between the two points 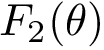
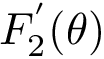
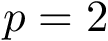
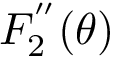
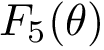
This distance is also directly expressed using the angles:
![\[d(X(\theta), X(\theta_i)) = \min( \fabs{\theta-\theta_i}, 2\pi - \fabs{\theta-\theta_i}).
\]](form_508.png)
For an integer 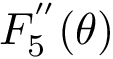
![\[\label{eq:Fp}
F_p(\theta) = \sum_{i=1,\dots,n} f_i(\theta), \text{ with } f_i(\theta)=d^p(X(\theta), X(\theta_i)).
\]](form_510.png)
Consider the minimum of this function, that is
![\[\label{eq:Fp-min}
\theta^* = \arg\min_{\theta\in[0,2\pi)} F_p(\theta).
\]](form_511.png)
The value 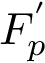
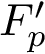
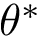
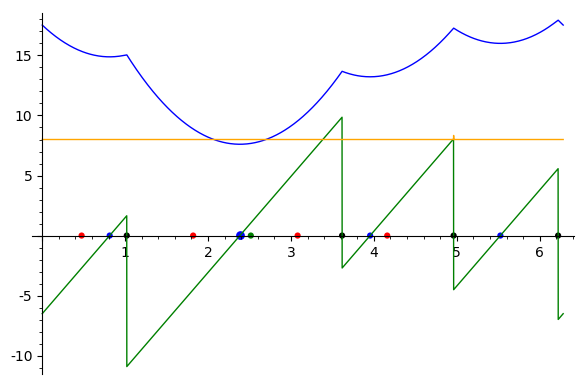 |
Sum of square distances function Sample points are in red, their antipodal points on the |
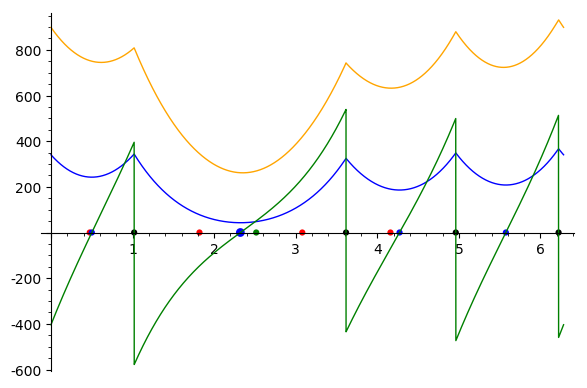 |
| Sum of distances to the power 5 function Sample points are in red, their antipodal points on the |
In short, the algorithm [50] subdivides the unit circle into interval namely circle arcs defined by data points and their antipodal values. This set of angles is denoted 
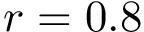
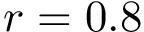


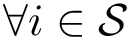
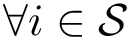
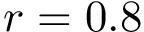
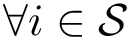

Albeit simple, the algorithm is numerically challenging since it deals with piecewise polynomials with transcendental coefficients (involving 
Following the classical design pattern in computational geometry, this package provides a generic implementation, templated by a kernel providing number types and associated predicates and constructions.
The main class is SBL::GT::T_Frechet_mean_S1.
Two kernels are provided:
Note that in our case, an erroneous evaluation of predicates may yield the loss or erroneous identification of a local minimum.
This package provides two executables:
This corresponds to the two above mentionned kernels.
See the following jupyter notebook:
import re #regular expressions
import sys #misc system
import os
import shutil
#import shutil # python 3 only
#input filename and output directory
odir = "results"
if not os.path.exists(odir):
os.system("mkdir results")
#choose value of p
p=4
angles_file="data/angles.txt" #file containing angles
weights_file="data/weights.txt" #file containing weights
exe_file="sbl-Frechet-mean-using-real-value-NT.exe" #App to compute minimas (sbl-Frechet-mean-using-interval-NT.exe is another option)
# run command(-r is used for specifiying radii input data -a to get all minimas)
cmd = "./%s -f %s -w %s -r -p %d -a -o %s" % (exe_file,angles_file,weights_file,p,odir)
os.system(cmd)
# load visualisation script and instanciate using data and corresponding minimas
minimas_file="results/angles_local_minima.txt"
load("centroid-circle.sage")
algo = Project()
algo.load_angles(angles_file,False)
algo.load_weights(weights_file)
algo.load_minimas(minimas_file,False)
algo.run(p)
#change value of p
p=5
# rerun command(-r is used for specifiying radii input data)
cmd = "./%s -f %s -w %s -r -p %d -a -o %s" % (exe_file,angles_file,weights_file,p,odir)
algo.run(p)