 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
Authors: F. Cazals and A. Chevallier and T. Dreyfus
This package defines generic functors to compute the potential energy 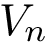
Atomic and coarse grain representations are supported.
The potential energy decomposes as a sum of contributions, which themselves decomposes into terms. The functors give access to contributions and terms.
Instantiations of these generic functors are provided for several classical force fields, including CHARMM, Amber, Martini. Additional force fields are easily incorporated using the FFXML format, from the OpenMM library.
Novel force fields are also easily defined by defining terms, which add up to define contributions.
The ability to support various force fields aims at leveraging three key operations on energy landscapes, namely:
exploring energy landscapes, see package Landscape_explorer
analyzing energy landscapes, see package Energy_landscape_analysis
NB: The computation of potential energy gradients (usually used as forces) using on automatic differentiation tools, will be provided elsewhere.
To describe the potential energy of a macro-molecular system, we shall use the following terminology:
particle class (for atoms or pseudo-atoms) : a class of particles. Example for proteins: C-alpha (CA) carbons.
To describe a force field, we shall need:
(bonded terms) pairs / triples / quadruples of classes of particle types;
The following examples stress the roles of particle types versus particle classes.
A typical expression for 
Bonded contributions: consist of three terms: bond / valence / torsion terms. These terms are best described in terms of internal coordinates.
Non bonded contribution: typically consists of van der Waals and electrostatic terms.
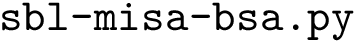
with
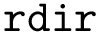
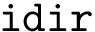
The number of parameters used to compute these six contributions is provided in a 6-tuple denoted 
The following remarks are in order:
Covalent terms are expressed with primitive internal coordinates (Molecular_coordinates), while non-covalent terms are expressed with pairwise distances.
The 6-tuple aims at scaling the complexity of a force field, which depends on the different possible combinations of particle types in a force field. See Examples: instantiating classical force fields .
![$ U_{\text{total}} = \sum_{\text{bonds}} K_r(r - r_{eq})^2 + \sum_{\text{angles}}
K_\theta(\theta - \theta_{eq})^2 + \sum_{\text{dihedrals}}
\frac{V_n}{2}\left[1 + \cos(n\phi - \gamma)\right] + \sum_{i \leq
j}\left[\frac{A_{ij}}{R_{ij}^{12}} - \frac{B_{ij}}{R_{ij}^6} +
\frac{q_iq_j}{\epsilon R_{ij}}\right]
$](form_889.png)
The following units are used consistently:
Mechanisms to ensure the compile time coherence of expressions involving such units are provided in the package UnitSystemTraits .
The potential energy 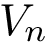
There is a unique class provided for computing the harmonic bond length term that is SBL::CSB::T_Molecular_potential_energy_bond_length_term_harmonic . For two bonded particles, the formula is :
where 
Two classes are provided to compute the harmonic bond angle term :
1. Harmonic valence angle term : SBL::CSB::T_Molecular_potential_energy_bond_angle_term_harmonic . For three bonded particles, the formula is :
with 
2. Urey Bradley term : SBL::CSB::T_Molecular_potential_energy_bond_angle_term_Urey_Bradley . For three bonded particles, the formula is :
with 
There are four classes provided for computing the possible torsion angle terms :
1. Harmonic torsion angle term : SBL::CSB::T_Molecular_potential_energy_torsion_angle_term_harmonic . For four bonded particles, the formula is :
where 
2. Periodic torsion angle term : SBL::CSB::T_Molecular_potential_energy_torsion_angle_term_periodic . For four bonded particles, the formula is :
where 

3. Fourier torsion angle term : SBL::CSB::T_Molecular_potential_energy_torsion_angle_term_Fourier . For four bonded particles, the formula is :
where 
4. Secondary structure knowledge based torsion angle terms: in selected coarse grain models, where the covalent structure is a simple sequence of pseudo-atoms : SBL::CSB::T_Molecular_potential_energy_torsion_angle_in_linear_structure_term . For four successive particles, the formula is :
where 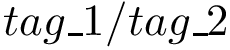
Improper dihedral angles are defined in Molecular_coordinates. Such angles do not correspond to a physical interaction between the four atoms involved; instead, they are used to enforce planarity constraints e.g. in cycles.
The difficulty with such angles is the lack of a standard. To see why, recall that for a given central atom sharing bonds to three neighbors, there are three different improper angles–see Molecular_coordinates. To make the exploitation of improper angles canonical, we proceed as follows, the rationale being to enforce geometric planarity:
There are two classes provided for computing the different non bonded terms :
1. To compute the vdW term : SBL::CSB::T_Molecular_potential_energy_Lennard_Jones_term . For two non bonded particles, the formula is :
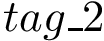
where 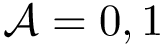
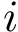
To compute a simplified electrostatic term : SBL::CSB::T_Molecular_potential_energy_electrostatic_term . For two non bonded particles, the formula is :
where 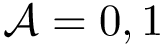
Non protein molecules as well as atoms (ions) can be added into the covalent structure defined in the package Molecular_covalent_structure. This can be done in two ways:
at the application level, by listing the molecules in the file describing the molecular system of interest.
We use the FFXML format to represent force fields.
In order to generate the FFXML files, it is necessary to convert the various existing formats with different scripts. For example, Amber and CHARMM have different representations of the force fields. In addition, depending on the molecule to study (sugar, peptide, protein, ...), and in which conditions they are presented (e.g, with or without solvant) , the parameters to load will be different.
To do so, we first resort on existing scripts if any, or define our own scripts otherwise.
The FFXML format (for Force Field XML format) is a simplified and hierarchical format for representing a given force field. It can be manually written for simple force fields as in section BLN69 , or it can be automatically created using the OpenMM library or the SBL scripts. For each case, the procedure for creating such an XML file is described in its own section.
The global architecture of a FFXML file is as follows :
<ForceField> <AtomTypes> </AtomTypes> <Residues> </Residues> <HarmonicBondForce> </HarmonicBondForce> <HarmonicAngleForce> </HarmonicAngleForce> <PeriodicTorsionForce> </PeriodicTorsionForce> <NonbondedForce coulomb14scale="1.0" lj14scale="1.0"> </NonbondedForce> </ForceField>
Depending on the input force field, some tags may appear or disappear. For example, an Amber force field is represented here in the FFXML format : Amber in FFXML.
reading a rational value is possible since our loader uses CGAL rational number type;
the previous rational value is converted to a floating-point representation since calculations whose result 
The OpenMM library provides a Python script processAmberForceField.py for generating FFXML files from Amber files. Generating a FFXML file for amino acids is simply done with the following command :
processAmberForceField.py amber16/dat/leap/lib/all_amino03.lib amber16/dat/leap/lib/all_aminoct03.lib amber16/dat/leap/lib/all_aminont03.lib amber16/dat/leap/parm/parm10.dat > amber16.ffxml
The first three files describe the topology of the different amino acids of a peptidic chain, in the middle, at the C-terminus and at the N-terminus. The last file describes all the bonded / non-bonded parameters.
The SBL provides the Python script sbl-process-CHARMM-force-field.py based on the ParmEd library. As for Amber, it loads CHARMM parameter files and generates a FFXML file based on the input parameters :
sbl-process-CHARMM-force-field.py toppar/top_all36_prot.rtf toppar/par_all36_prot.prm
The first file describes the topology of the different amino acids of a peptidic chain (at any position). The last file describes all the bonded / non-bonded parameters.
The Martini force field is provided in the Gromacs format. The SBL provides the Python script sbl-process-Martini-force-field.py for processing Martini force fields in such a format. It takes as input a itp file defining the topology and the bonded / non-bonded parameters :
sbl-process-Martini-force-field.py martini_v2.2P_aminoacids.itp
The number of parameters being very small, and uniquely identified, we do not provide any script for generating the corresponding FFXML. We rather provide the corresponding FFXML file : BLN69 Force Field
The following examples computes the potential energy with different force fields, following the same pattern :
Amber refers to a suite of force fields, see Amber. We consider here the ff14sb force field from Amber. The 6-tuples of this force filed are:
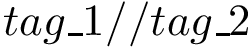
Note that there is no Lennard-Jones contribution in this force field. Note also that the charge is defined separately for each particle type, explaining the high number of parameters for the electrostatic contribution.
The ffxml file of the ff14sb force field is given here : ff14sb Amber Force Field.
The following example instantiates the Amber potential energy using the included file Force_field_atomic.hpp (see section Advanced : defining potential energy types for the specifications) :
CHARMM refers to a suite of force fields, see CHARMM. We consider here the charmm36 force field for proteins. The 6-tuples of this force filed are
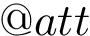
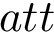
The ffxml file of the charmm36 force field is given here : CHARMM Force Field.
Note that the example of section Amber works also for this type of force field since the data structures are exactly the same.
The Martini force field is a coarse grain force field designed to be used with Gromacs . The 6-tuples of this force filed are
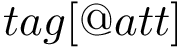
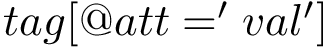
The ffxml file of the Martini force field is given here : Martini Force Field. In order to use the Martini force field in the SBL, two steps are required:
The following example instantiates the Martini potential energy :
BLN69 is a protein model based on three types of particles, each representing a pseudo amino-acid – see Example Landscapes defined from mathematical functions . There is a specific force field based on the coarse grain model of BLN69. The 6-tuples of this force filed are
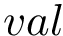
Note that there is a unique set of parameters for bonds and for angles. Note also that neither dihedral nor electrostatic terms are present in this force field.
See the BLN69 Force Field. See Example Landscapes defined from mathematical functions for more details about BLN69.
The following example instantiates the BLN69 potential energy :
This section introduces the different types defining the potential energy, used in the examples above.
In the sequel, we take for granted the enumeration of primitive internal coordinates, as explained in the package Molecular_coordinates.
Three important types of C++ data structure are defined in this package. Following definition fig-eq-ff, contributions are defined from terms which are themselves defined from visitors:
Visitor : a visitor is a class defining an iterator over a collection of entities, entities being pairs, triples or quadruples of particles. An entity can represent an non-bonded pair of particles, a bond, a valence angle or a torsion angle. Visitor provided by the SBL are templated by the type CovalentStructure representing the type of the covalent structure (see package Molecular_covalent_structure). It defines the type Iterator representing the iterator itself, and the static methods begin( CovalentStructure ) and end( CovalentStructure ) for starting/ending the iteration over the target entities.
Example : iterating over all bonds is done using the class SBL::CSB::T_Bonded_particles_visitor< CovalentStructure >.
Term : a term is a functor computing one term of the sum of a contribution of 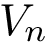
Contribution : a contribution is a functor computing the contribution of 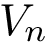
Instantiating a potential energy function consists then in defining different contributions, and to sum them. For example, the class SBL::CSB::T_Molecular_potential_energy_bonded_contribution instantiates the three contributions related to the bond distances, bond angles and torsion angles, and sum them up. It is possible to instantiate in addition the Lennard Jones contribution from the classes SBL::CSB::T_Molecular_potential_energy_Lennard_Jones_term and SBL::CSB::T_Unbonded_particles_visitor for computing a basic potential energy without electrostatic term. Adding the electrostatic contribution is done in the same way by using the class SBL::CSB::T_Molecular_potential_energy_electrostatic_term
typedef SBL::CSB::T_Bonded_particles_visitor<Covalent_structure> Visitor;
typedef SBL::CSB::T_Molecular_potential_energy_bond_length_term<Conformation,
Covalent_structure,
Parameters> Term;
typedef SBL::CSB::T_Molecular_potential_energy_term<Visitor, Term> Term;
Conformation C;
Covalent_structure S;
Contribution contrib;
double e = contrib(C, S);
typedef SBL::CSB::T_Unbonded_particles_visitor<Covalent_structure> Visitor;
typedef SBL::CSB::T_Molecular_potential_energy_Lennard_Jones_term<Conformation,
Covalent_structure,
Parameters> Term;
typedef SBL::CSB::T_Molecular_potential_energy_contribution<Visitor, Expression> Contribution;
Conformation C;
Covalent_structure S;
Contribution contrib;
double e = contrib(C, S);
We need to define the types to represent a conformation, and to load conformations from an input file.
#include <SBL/Models/Geometric_conformation_traits.hpp> #include <SBL/Models/Conformations_file_loader.hpp> typedef SBL::Models::T_Geometric_conformation_traits<> Conformation_traits; typedef Conformation_traits::Conformation Conformation; typedef SBL::Models::T_Conformations_file_loader<Conformation> Conformations_loader;
Four types have to be defined :
#include <SBL/CSB/Particle_info_proteins.hpp> #include <SBL/CSB/Molecular_covalent_structure.hpp> #include <SBL/IO/Molecular_covalent_structure_loader_from_PDB.hpp> typedef SBL::CSB::T_Particle_info_for_proteins<> Particle_info; typedef SBL::CSB::T_Molecular_covalent_structure<Particle_info> Covalent_structure_base; typedef SBL::CSB::T_Molecular_covalent_structure_builder_for_proteins<Covalent_structure_base> Covalent_structure_builder; typedef SBL::IO::T_Molecular_covalent_structure_loader_from_PDB<Covalent_structure_base, Covalent_structure_builder> Covalent_structure_loader;
All the parameters used to compute the potential energy function are gathered in the class SBL::IO::T_Potential_energy_parameters< CovalentStructure , UnitSystemTraits > : these parameters corespond to the force field parameters, and to the units used to represent the constants in the force field parameters. The parameter CovalentStructure is a covalent structure as defined in the package Molecular_covalent_structure, and the parameter UnitSystemTraits defines the unit system to be used, as defined in the package UnitSystemTraits.
This class also provides a loader for the ffxml files, and gives a naive acces to the corresponding parameters. Instantiating this class can be done simply like this :
#include <SBL/IO/Potential_energy_parameters.hpp> #include <SBL/Models/Unit_system_traits_none_AKMA.hpp> #include <SBL/Models/Unit_system_traits_for_potential_energy.hpp> #include <SBL/CSB/Molecular_potential_energy_structure_parameters_traits_default.hpp> typedef SBL::Models::T_Unit_system_traits_none_AKMA<double> UST_base; typedef SBL::Models::T_Unit_system_traits_for_potential_energy<UST_base> UST; typedef SBL::IO::T_Potential_energy_parameters<Covalent_structure_base, UST> Potential_energy_parameters_loader;
The way parameters are built and the access to the covalent structure data structure can be time consuming, in particular in the context of computing potential energy and its gradient. To circumvent this difficulty, it is possible to use optimized data structures, with less functionality but more efficient access :
Once the base covalent structure and the base parameters have been loaded, it is possible to build a new covalent structure object and a new parameters object with constant time access to each parameter, and with the possibility to visit only entities (particles bonds or angles) having a non-zero contribution to the potential energy.
In order to do so, the class SBL::CSB::T_Molecular_potential_energy_structure_parameters_traits_optimal< CovalentStructureBase , PotentialEnergyParametersBase > provides three important types :
In this way, the following code allows to simply define the optimal versions :
#include <SBL/CSB/Molecular_potential_energy_structure_parameters_traits_optimal.hpp> typedef SBL::CSB::T_Molecular_potential_energy_structure_parameters_traits_optimal <Covalent_structure_base, Potential_energy_parameters_loader> Structure_parameters_traits; typedef Structure_parameters_traits::Covalent_structure Covalent_structure; typedef Structure_parameters_traits::Potential_energy_parameters Potential_energy_parameters; typedef Structure_parameters_traits::Build_structure_parameters Build_structure_parameters;
In order to minimalize the code changes to switch from one representation to the other, the class SBL::CSB::T_Molecular_potential_energy_structure_parameters_traits_default is also provided :
#include <SBL/CSB/Molecular_potential_energy_structure_parameters_traits_default.hpp> typedef SBL::CSB::T_Molecular_potential_energy_structure_parameters_traits_default <Covalent_structure_base, Potential_energy_parameters_loader> Structure_parameters_traits; typedef Structure_parameters_traits::Covalent_structure Covalent_structure; typedef Structure_parameters_traits::Potential_energy_parameters Potential_energy_parameters; typedef Structure_parameters_traits::Build_structure_parameters Build_structure_parameters;
Two important types have to be defined :
#include <SBL/CSB/Molecular_potential_energy.hpp> typedef SBL::CSB::T_Molecular_potential_energy_bonded_contribution <Conformation, Covalent_structure, Potential_energy_parameters> Bonded_contributions; typedef SBL::CSB::T_Unbonded_particles_visitor<Covalent_structure> Non_bonded_visitor; typedef SBL::CSB::T_Molecular_potential_energy_Lennard_Jones_term <Conformation, Covalent_structure, Potential_energy_parameters> Lennard_Jones_term; typedef SBL::CSB::T_Molecular_potential_energy_electrostatic_term <Conformation, Covalent_structure, Potential_energy_parameters> Electrostatic_term; typedef SBL::CSB::T_Molecular_potential_energy_contribution <Non_bonded_visitor, Lennard_Jones_term, Electrostatic_term> Non_bonded_contributions;
We have seen above that adding up terms of a given types makes up a contribution. Sometimes, it is useful to inspect closely all terms of a given type. As an example, energy type refers to the Lennard Jones potential in the sequel.
We now describe the mechanism implemented to dump all such terms into a file, in four steps.
First, the class SBL::CSB::T_Molecular_potential_energy_term_static_info defines a static variable indicating that all terms of a given energy type must be dumped into a file, and the corresponding filestream. Note that the class is templated by a TermType: we indeed want the aforementioned variables to exist for this specific energy type only.
Second, the term of interest, SBL::CSB::T_Molecular_potential_energy_Lennard_Jones_term, in our case, inherits from this static class. To instantiate the class SBL::CSB::T_Molecular_potential_energy_term_static_info , we use the Curiously recurring template pattern.
Third, the functor of the energy type, SBL::CSB::T_Molecular_potential_energy_Lennard_Jones_term , does the energy computation and if the flag is set, dumps the result into a file.
Fourth, the flags and the output file name are set in the main application, as static members:
Lennard_Jones_term::s_dump_flag = true; Lennard_Jones_term::s_ofname = "/tmp/LJ-terms.txt"; Lennard_Jones_term::s_ostream;
Naturally, the values of these static variables can be set using boost::program_options. As an example, checkout sbl-energy-charmm.{cpp,exe}. The following call generate the file /tmp/LJ-terms.txt containing LJ terms for all non bonded pairs:
sbl-energy-charmm.exe -c ala5.txt -s ala5.pdb -p charm.xml --energy-terms
For each potential energy term, this package provides also an associated gradient term. Recall that potential energy terms are expressed with internal coordinates, while the input is expressed with cartesian coordinates. Therefore a potential energy term 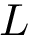
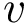
This package provides gradients for the functions 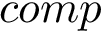
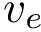
A numerical estimate using finite differences is also provided with the class SBL::CSB::T_Molecular_potential_energy_numerical_gradient, allowing to verify the accuracy of the computed gradients.