 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|

Authors: F. Cazals
Consider a set of clusters, each given in the form on a 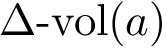

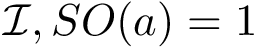
This package provides a number of geometric analysis on clusters:
The distance to measure of a point 

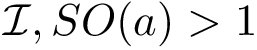
The DTM transform of the point set 
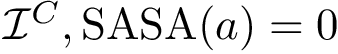
The DTM is very useful to detect outliers and denoise the dataset. Indeed, in the presence of outliers, a first mode is observed for typical distances between an inlier and its neighbors, while larger (and less pronounced) modes are observed for outliers.
Parameter(s). The number of neighbors. Practically, one uses 
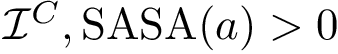
PCA scree plots. Upon centering the data matrix 
The scree plot is the line plot of the eigenvalues of PCA, by decreasing contribution.
The covariance dimension is the minimum number of directions required to explain a predefined fraction of the variance [186] .
Parameter(s). The fraction of explained variance which determines the covariance dimension.
Once the covariance dimension of each cluster has been determined, a proxy for the cluster is given by its center of mass and the vector space spanned by the principal vectors selected.
Principal angles: definition. Consider two affine spaces 
The 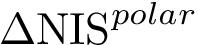
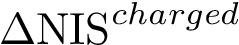
with
under the constraints
Assume the affine spaces are given by two matrices 
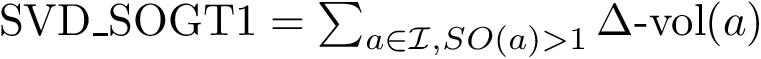
Consider the SVD of matrix 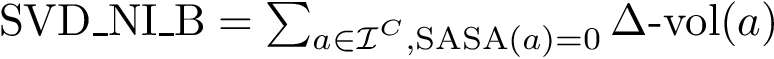
![\begin{equation}\begin{cases}
[u_1,\dots, u_p ] = Q_F U,\\
[v_1,\dots, v_q ] = Q_G V,\\
\cos \theta_k = \sigma_k, k:1,\dots,q.
\end{cases}
\end{equation}](form_89.png)
If the two affine spaces are given by matrices of rank 
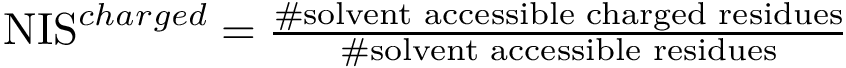
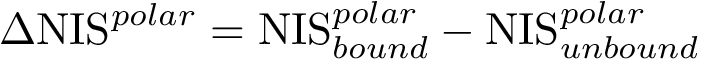

Principal angles: distances. One can therefore compare two clusters using the following distances between subspaces [197] :

Following ye2016schubert , it is convenient to distinguish:
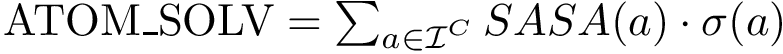
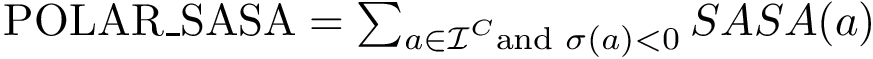
The Hausdorff affine distance. For a point 
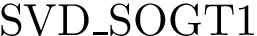
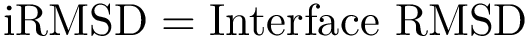
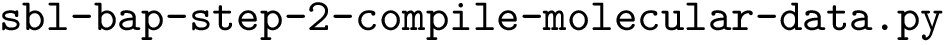
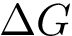
We define the one-sided Hausdorff affine distance between the point set 
Mimicking the Hausdorff distance, we define the Hausdorff affine distance between 
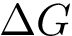

To compute the distance from 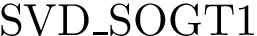

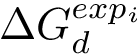
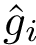
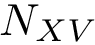
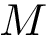
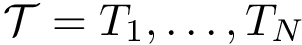
The Average/mean Hausdorff affine distance. In case of outliers, the previous distance put emphasis on the maximum distances.
To tame down this difficulty, define the average one-sided Hausdorff affine distance between the point set 
![\begin{equation}\distMHAOS{B}{P_a} = \bigl( \frac{1}{\size{P_a}} \sum_{x\in P_A} \distproj[p]{B}{x} \bigr)^{1/p}.
\end{equation}](form_110.png)
Mimicking the Hausdorff distance, we define the average/mean Hausdorff affine distance between 
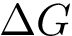
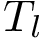
 |
| Distance from a point |
Parameter(s). The dimension of the vector spaces compared is determined by the fraction of explained variance used to determine the covariance dimension.
SESC. The following is developed in [goldenberg2024subspace] .
Given a point set in 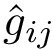
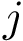
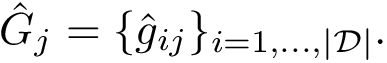
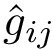
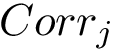
Parameter(s). Two hyperparameters:
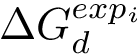
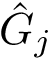
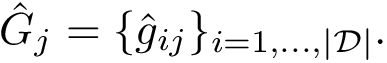
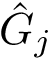
Associated Bayesian Information Criterion – BIC. Consider a cluster of dimension 
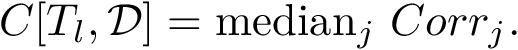
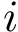
Support vector machines (SVM) is a classical tool to perform classification [174] . Of particular interest is the maximum margin separation , that is the width of the slab separating two point sets. Computing this margin boils down to a quadratic problem. Misclassified items can be tolerated, using a so-called regularization of the optimization problem.
Given a point set, a natural question is to ask whether this point set is naturally separable . We provide this insight in two steps:
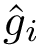
We summarize with the gap/span ratio , namely the ratio between the maximum margin width, and the extent of the projection of all samples onto the line perpendicular to the separating hyperplanes.
Parameter(s). The regularization coefficient 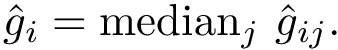
Notions of high dimensional medians generalizing the univariate median are useful concepts to obtain a notion of center point of a point set, see [123], [195] and references therein.
Various such generalizations have been proposed, in including the Fermat-Weber point which minimizes the sum of Euclidean distances to data points, the Tukey median defined as any point whose Tukey depth is at least 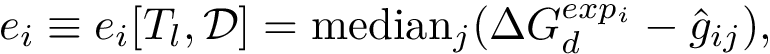
The projection median is defined by projecting the dataset onto random lines, computing the univariate median for each projection, and computing a weighted average of the data points responsible for these univariate medians, see [82] and [18] . It is an elegant, stable and remarkably effective generalization of the univariate median. This package provides an implementation of the projection median as a weighted sum of the input points [83] .
This package provides the script sbl-cluster-analysis.py, whose input(s) and output(s) are as follows.
Input:
Output:
We use the PCA class from SciKitLearn. The following points are worth noticing:
Covariance dimension. Selection of the covariance dimension as a function of the explained variance:
pca = PCA(n_components = explained_var_target_value, svd_solver='full')
Note that the principal directions are accessed via pca.components_
Projection onto affine space. Projection of a point 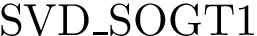
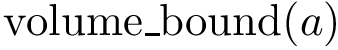
point = X[i,:] # flat array (d,)
if Y_com is None: # vector mode
vec = point
else: # affine mode
vec = point - Y_com
# Project the vector onto the affine space. with d1 the dim of the vector space
# (d,d1)x(d1,d)x(d,1) yields a (d,) flat array: pt in the ambient space
v_proj = Y_obasis @ (Y_obasis.T @ vec.T) # flat array (d,)
v_ortho = vec-v_proj
# Compute the distance
d = np.linalg.norm(v_ortho)
The previous analysis are implemented in the following modules of the package Clustering_analysis: Point_cloud_DTM_filtering, Principal_angles, SVM_cluster_challenger.
The dependencies are as follows: