 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|

Authors: F. Cazals and C. Robert
As its title figure suggests, this package makes it possible to coherently handle proteins and nucleic acids. As such, it has strong connexions with the following packages:
The documentation of this package is kept as concise as possible, and we only focus on the architecture used to handle both types of biomolecules.
The following snippet (from sbl-biomolecule-info.cpp) shows how to load a file and access the corresponding biomolecules and their chains (protein: polypeptide chains; nucleic acid: polynucleotide chains).
File: sbl-biomolecule-info.cpp
This section presents the architecture for handling biomolecules.
To facilitate reading, recall that a builder builds a data structure, while a loader loads a PDB/mmCIF file and calls a builder.
In short, the construction of biomolecules from a PDB/mmCIF file is handled as follows:
Step 0. The molecular system is loaded from the PDB/mmCIF file.
Step 1.
A unique molecular covalent structure is built, in two steps: first, the residues of the molecular systems are linked into polymer chains of amino acids or nucleotides; second, each polymer chain is added to the MCS.
In general, polypeptide and polynucleotide chains are disconnected. Which means that upon building all polymers, the number of connected components of the MCS is exactly the number of chains.
We now detail these steps and the associated classes.
![\begin{algorithm}\begin{algorithmic}[1]
\Procedure{BiomolecularRepresentationLoader}{PDB/mmCIF file(s)}
\State Load file(s) and record sequence of residues and conformations
\For{each file}
\State Call MCS builder to build the global MCS of that file
\State Extract the protein and nucleic acid representation
\EndFor
\EndProcedure
\end{algorithmic}
\end{algorithm}](form_38.png)
We now detail the previous steps, referring to the C++ classes provided.
Hierarchy of MCS builders. As noticed above, a MCS builder creates a graph with as many subgraphs as there are chains in the biomolecule. Because our builders handle linear polymers, they are implemented in a generic fashion which factors out common functionalities – Fig. fig-builders-diamond.
We have seen in the Molecular_covalent_structure package that various instantiations of MCS are available, corresponding to the pieces of information associated with an atom. As detailed in section Molecular Covalent Structure: instantiations, the corresponding suffixes are FIT, FIAT, HIT, and HIAT.
 |
| Molecular covalent structure builders: the diamond pattern used for biomolecules. The generic builder handles operations common to all linear polymers. The two derived classes handle chains of amino acids and nucleotides, respectively. Finally, the builder for biomolecules uses the previous two as template parameters, to convert connected stretches of residues into a molecular covalent structure. |
The MCS builder for biomolecules and the switch Protein vs Nucleic Acid. A MCS builder for biomolecules requires template parameters specifying a builder for proteins and one for nucleic acids. Given these template parameters, the sketch of the construction, using a switch to distinguish proteins from nucleic acids, is as follows:
File: Molecular_covalent_structure_builder_biomolecules.hpp
The following are critical observations:
In the sequel, builder refers to an instance of this builder for biomolecules.
Step 1: building the global MCS with the MCS loader. The MCS loader loads PDB files and call the builder building one MCS per PDB/mmCIF file processed.
File: Molecular_covalent_structure_loader.hpp
Step 2: building the protein and nucleic acid representations with the Biomolecule representation loader.
The biomolecule representation loader is parameterized by the two returned types (ProteinRepresentation and NucleicAcidRepresentation), and the class used to perform Step 1 (the MCSLoader).
File: Biomolecule_representation_loader.hpp
The instantiation using the aforementioned lineage HIT of data structures is:
Then, the creation of protein and nucleic acid representations goes as follows:
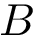
File Biomolecule_representation_loader.hpp
NB: In the code above, get_my_chain_type() refers to the fact that the MCS used to create a protein representation necessarily hosts a single chain, as opposed to the global MCS used in Step 1.