 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
Authors: S. Guo and E. Sarti and F. Cazals 
The Plugin_manager package enables the automatic creation of cross‑platform graphical user‑interface (GUI) plugins for command‑line applications, from a single specification. Its objectives are to:
In theory, developing GUI for 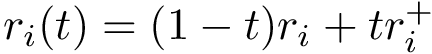
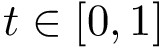
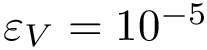
We instead propose a solution requiring in 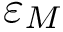
 |
| Plugin generation using one specification file per application. For each application |
A number of plugins are available, see List of available plugins .
To regenerate the GUIs for all of the above at once, run the orchestration script (this is not needed to run because we have already generated them):
Refer to section Installing and using SBL with Pixi (Linux 64, OSX 64, OSX ARM64) in the SBL Installation Guide for detailed steps on creating a Pixi environment, installing the SBL package, and running sbl-install-plugins.
Once SBL and the plugins are installed, you can use them as follows:
VMD plugins
Open VMD and navigate to Extensions → SBL to access the available plugins.
PyMOL plugins
Open PyMOL and navigate to Plugin menu to access the installed SBL plugins.
Web plugins
Launch the Web plugins dashboard:
Then open the URL printed in the terminal (typically http://127.0.0.1:8700/) in your web browser.
If you prefer to install a single plugin independently (without the full SBL installation), follow these platform‑specific instructions.
Installing an individual VMD plugin
sbl_vorlume_pdb) containing pkgIndex.tcl and sbl_pluginname.tcl. sbl_pluginname.tcl to match your plugin (e.g., sbl_vorlume_pdb.tcl) and update all occurrences of pluginname inside both files with the actual plugin name. ~/sblvmdplugins is in VMD's auto_path. Add to ~/.vmdrc if needed: ~/.vmdrc: ~/.vmdrc (required for plugin communication). Skip this step if the configuration is already present: vmd_visualize_sbl_plugin convenience proc to ~/.vmdrc (for loading SBL visualization scripts). Skip this step if the proc is already defined: Installing an individual Web plugin
http://127.0.0.1:8700/.Installing an individual PyMOL plugin
PyMOL plugins using pymol.Qt can be installed directly via PyMOL's plugin system:
sbl_vorlume_pdb) containing __init__.py and gui.py. Alternatively, use PyMOL's built‑in Plugin Manager:
__init__.py file.
Plugin: structure. A generated plugin is organized into four areas (Fig. GUI architecture):
We adopt the Model–View–Presenter (MVP) pattern for both the generator and the generated plugins:
Plugin at work: data flow.
On desktop platforms, VMD and PyMOL execute the viewer scripts (.vmd or .py) produced by the post analysis script and render them in their display windows. In the web platform, a Panel application embeds or links to an NGLViewer / Three.js viewer page served on a standalone port. The viewer is refreshed based on the json file generated by the post‑analysis script.
 |
| GUI: generic architecture A plugin consists of four main areas: (1) the Input area enabling the specification of input data, (2) The output are to display statistics and figures, (3) The 3D molecular graphics area, and (4) The update area for refreshing outputs. |
Plugin design: the three steps. A typical workflow comprises three phases:
The user designed associated files are as follows:
layout.ui — Qt Designer file defining geometry for input/output/update areas.selected_flags.txt — plain‑text list of CLI flags to expose in the input area.update_area_flags.txt — optional plain‑text list of flags for the post‑analysis script.post_analysis.py — Python script that converts raw CLI outputs into panel‑ready and viewer-ready files.
The script run_generator.sh A convenience script (run_generator.sh) in Applications/ <AppName>/plugins automates code generation. The command: bash run_generator.sh, which run in the plugins folder will produce the three GUIs: Tkinter for VMD, PyQt6 for PyMOL, and Panel for the web under the folder Applications/ <AppName>/plugins/step3_generated_code. Fig. GUI design workflow shows the design workflow of SBL GUI plugins using an example.
Files generated. For each application, say SBL_DIR/Application_XXX, the 
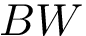
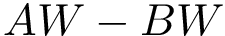
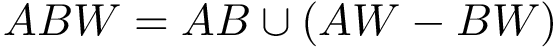
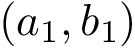
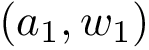
 |
| GUI: design example An example of using the sbl-gui-generator.py to automatic create cross‑platform graphical user‑interface (GUI) plugins for command‑line applications. |
Step 1: Preparing layout.ui. Requirements:
inputPanel. The update area is optional and must be named updatePanel.outputText\d *, outputFigure\d *, outputPDF\d *, outputTable\d *, outputHtml\d *, or outputHtmlTabs.This .ui file is created using Qt Designer and saved as layout.ui.
The following is the layout file for the plugin sbl_spectraldom from the package Spectral_domain_explorer :
Step 1: Preparing selected_flags.txt. This file lists the options to expose in the inputPanel.
_) in flag names; use hyphens (-) instead.The following is the selected_flags.txt file for the plugin sbl_spectraldom from the package Spectral_domain_explorer-package:
Mapping to widgets:
Entry (file/directory options also provide a Browse Button) plus a tooltip Label (shows CLI help on hover).Checkbox plus a tooltip Label.Frame.Entry accepts extra CLI options.Run Button is appended, with a tooltip displaying the CLI header.Advanced usage of selected_flags.txt. Beyond the simple "one flag per line" format shown above, an optional CSV‑style format allows you to control tooltips, default values, and widget behaviour on a per‑flag basis. Each non‑comment line can contain up to four comma‑separated fields:
Blank fields fall back to the values parsed from the CLI --help output. Comment lines starting with # are ignored. An example is provided :
The four columns have the following meaning:
--help). --help (leave empty to use the help description). --help. For flags with an argument this fills the corresponding Entry; for boolean flags (no argument) this controls the initial state of the Checkbox (accepted truthy values include 1/true/yes/on/y/t; false values include 0/false/no/off/n/f). isInputStructure forces a file chooser to be attached to the Entry, and isOutputDir forces a directory chooser.
Step1: Preparing post_analysis.py. To align with widget names, the post‑analysis script must produce files using the following conventions:
outputFigure1 → outputFigure1.png (static image) and/or outputFigure1.dat (data for interactive figures)outputPDF1 → outputPDF1.pdf (PDF)outputTable1 → outputTable1.csv (table)outputText1 → outputText1.txt (text)outputHtml1 → outputHtml1.html (standalone HTML)1.vmd / 1.py / 1.json → viewer script or json for molecular graphics areaReference script for the SBL Intervor plugin: $SBL_DIR/Applications/Space_filling_model_interface/scripts/intervor_abw_atomic_plugin_post_analysis.py
Example call:
-d (executable output directory), -p (post‑analysis output directory), --post-flag <value from Update area, if present>, and --viewer <vmd|pymol|ngl> depending on the target platform. The plugin then refreshes its Output area and the molecular viewer based on the files produced by the post‑analysis script.The following is the plugin_post_analysis.py file for the plugin sbl_spectraldom from the package Spectral_domain_explorer-package:
Step1: Preparing update_area_flags.txt file. This optional file lists flags to expose in the updatePanel, which parameterize the post‑analysis step to update the outputs.
The following is the update_area_flags.txt file for the plugin sbl_spectraldom from the package Spectral_domain_explorer-package:
Step 2: Generating the plugin formal specification JSON file. Once layout.ui, intervor_abw_atomic_plugin_post_analysis.py, selected_flags.txt, and update_area_flags.txt are ready, generate platform‑specific code via running the bash script run_generator.sh in the plugins folder:
The above process call the main generator script sbl-gui-generator.py,
This first produces the formal specification JSON file intervor-ABW-atomic_gui.json, which will finally located in the folder Applications/ <AppName>/plugins/step2_formal_spec.
Step 3: Generating the plugin code. Then it produces the platform-specific plugin code based on the formal specification JSON file. This creates the three GUIs: VMD, PyMOL, and Web. The generated code is located under the folder Applications/ <AppName>/plugins/step3_generated_code as vmd.tar.gz, pymol.tar.gz, and web.tar.gz.
The used defined files layout.ui and selected_flags.txt and update_area_flags.txt are converted to a JSON specification that records widget types, geometry, and metadata.
In the following, we present the types and syntax used for this conversion.
The following widget types in JSON currently includes:
The JSON file consists of four sections–the first three being derived from the ui file:
Input panel section. Based on selected_flags.txt, we populate the children of inputPanel: flags with parameters from the CLI become an Entry (file/directory entries are paired with a Button to open a selector) followed by a tooltip Label that shows the help text on hover; boolean flags become a Checkbox plus a tooltip Label. Each flag’s controls are grouped in a Frame. An additional free–form Entry accepts extra CLI options. Finally, a Run Button is added with a tooltip showing the CLI header.
Output panel section. This section declares the widgets that present artifacts produced by the post-analysis step after the CLI finishes. Concretely, output panels are a set of typed widgets: TextArea, ImageView, TableView, each with a file pattern and refresh policy. After a run, the generator calls refresh_output_panel method: it scans output_dir for the newest files matching the source glob of each widget and updates the views accordingly.
Update panel section. This optional section declares widgets that control the post-analysis step. Based on post_analysis_flags.txt, these controls mirror input widgets but are not passed to the CLI; instead, they parameterize the post‑analysis. Changing any of these parameters and clicking the Update button triggers a re‑run of the post‑analysis and selectively refreshes the affected output widgets and/or the 3D view.
All output widgets are read‑only and refresh automatically after a successful run or when the Update area triggers a refresh.
Metadata section. Includes a metadata block (e.g., exe_name, post_script).
As already discussed, we automatically generated code for the targeted platform from the formal specification (Fig. fig-plugin-generation). The mapping between formal types and those used by the platforms is as follows:
 |
| Mapping between JSON widget types to types used by the backends. |
We detail the implement of the sbl-gui-generator method here.
The main entry point is 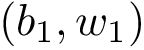
A convenience script, 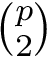
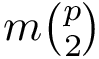
To generate a concrete plugin for an application, the generator copies the appropriate templates and injects application–specific pieces into 
 |
| Plugin manager modules. (A) Design phase. Using a layout editor, the designer (i) arranges the relative positions and sizes of widgets for the Input and Output areas; (ii) selects the CLI options (flags and arguments) to expose in the Input area; and (iii) specifies post–analysis parameters to surface in the optional Update area, whose changes selectively refresh the Output area and/or the 3D display. (B)Generation of the formal specification. A formal specification of the layout for the Input and Output areas is automatically generated. Plugin generation. (C) The python code of plugins is automatically generated. From the JSON, we synthesize frontend and backend code for the GUI libraries targeted (PyQt6, tkinter, or Panel). Widget types are mapped to their platform–specific counterparts and the generated backend logic constructs the CLI command, executes it, runs the post–analysis (if any), refreshes outputs, and displays results in the molecule viewer. |
The previously described workflow is implemented by the following modules (Fig. Code modules); each has a clear responsibility and can be combined to target different GUI frameworks:
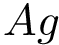
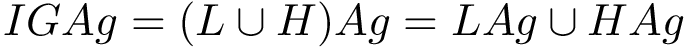
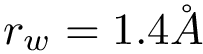
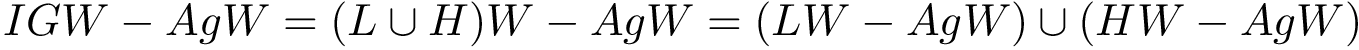
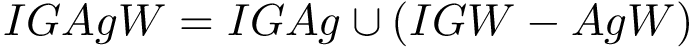
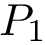
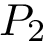
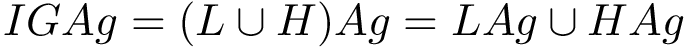
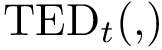
For PyQt6 and Tkinter, absolute positions and sizes from the 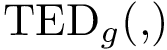
For Panel, which is row/column–based, we quantize absolute widget coordinates onto a fixed 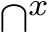
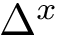
The grid size is 
A widget with bounding box 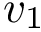
Indices are clamped to the grid bounds. An optional overlap resolver relocates later widgets (row‑major scan) to the first free slot that accommodates their span; if no slot exists, new rows are appended at the bottom. The final indices are stored in JSON for Panel code generation. Widths/heights smaller than one pixel are treated as 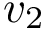
For PyQt6 and tkinter, absolute positions and sizes from the .ui file are used directly. For Panel, which is row/column–based, we assign grid coordinates from the absolute widget positions.
Templates root. Target–agnostic templates live under $SBL_DIR/Applications/Plugin_manager/template_files/ with three subfolders: vmd/, pymol/, and web-ngljs/, web-threejs.
VMD (Tkinter). Each VMD plugin template provides gui.py, pkgIndex.tcl, and sbl_pluginname.tcl, plus an mvp/ subtree with: config.py, presenter.py, services/, and ui/.
PyMOL (PyQt6). Same mvp/ structure as VMD (no Tcl files), with PyQt6–based UI code.
Web-NGL (Panel). Mirrors the same mvp/ layout and includes an ngl_viewer/ directory for NGL static assets.
Web-Three.js (Panel). Mirrors the same mvp/ layout and includes a threejs_viewer/ directory for Three.js static assets.
NGL viewer assets. The NGL static assets are developed in $SBL_DIR/Applications/Plugin_manager/viewer_app/ (TypeScript). Build them with:
The build outputs to $SBL_DIR/Applications/Plugin_manager/template_files/web/ngl_viewer/.
PANEL provides a unified framework to build dashboards and interactive applications in Python, leveraging the PyData ecosystem (e.g., Pandas, Matplotlib, Plotly). In our workflow, GUI-generation outputs are translated into Panel widgets and layout primitives. A typical launch command is:
While suitable for prototyping, this mode lacks production features such as HTTPS, traffic routing, process supervision and standard web port exposure. These limitations are addressed via our deployment stack.
Our Panel-based web GUIs embed advanced 3D viewers - for example NGL Viewer (molecular structures) or Three.js (geometry) — via iframes. Data (PDB/JSON) are exported by the post-analysis routine, the viewer is re-loaded with a cache-busting query parameter, and the rendering is handled entirely client-side. This separation of execution (server) and rendering (browser) simplifies deployment under web security policies. A schematic is shown in Fig. fig-panel-nglviewer-workflow .
 |
| Panel–NGL/Three.js integration workflow. The Panel-based web GUI embeds a dedicated viewer via an <iframe>; structural and geometric data are exported to JSON and PDB files, and the viewer is reloaded with a cache-busting query string to fetch updated content. |
Since the default panel serve app.py mode is intended for development rather than production, our deployment uses a robust production stack (Fig. fig-panel-deployment: the Panel app (via Bokeh server) is managed by the OS service manager (e.g., systemd), and sits behind Nginx which handles TLS termination and proxies HTTP/WebSocket traffic. Multiple apps can be hosted via URL prefixes.
 |
| Production deployment architecture for Panel apps. The Panel‐based web application (Bokeh server) listens on a local port (e.g., 5006) and is managed via systemd, while Nginx handles public access (ports 80/443), TLS termination, and forwards HTTP/WebSocket traffic to the app. |