 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
 |
Structural Bioinformatics Library
Template C++ / Python API for developping structural bioinformatics applications.
|
Packages | |
| ConformationMoveset | |
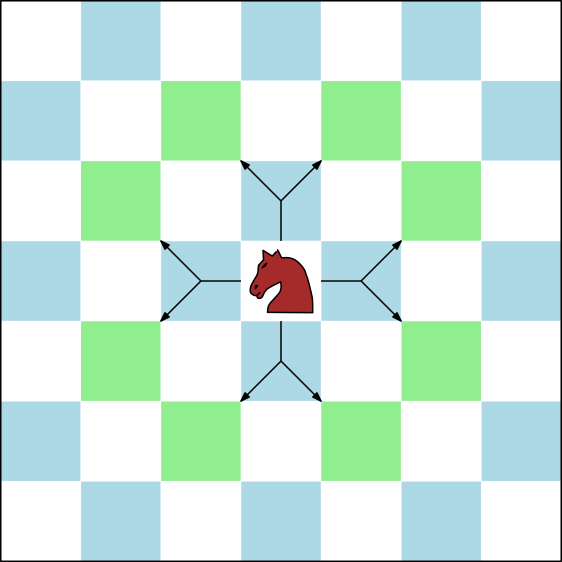 | This package provides different methods for extending a conformation to a new one Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus and C. Roth |
| MolecularGeometryLoader | |
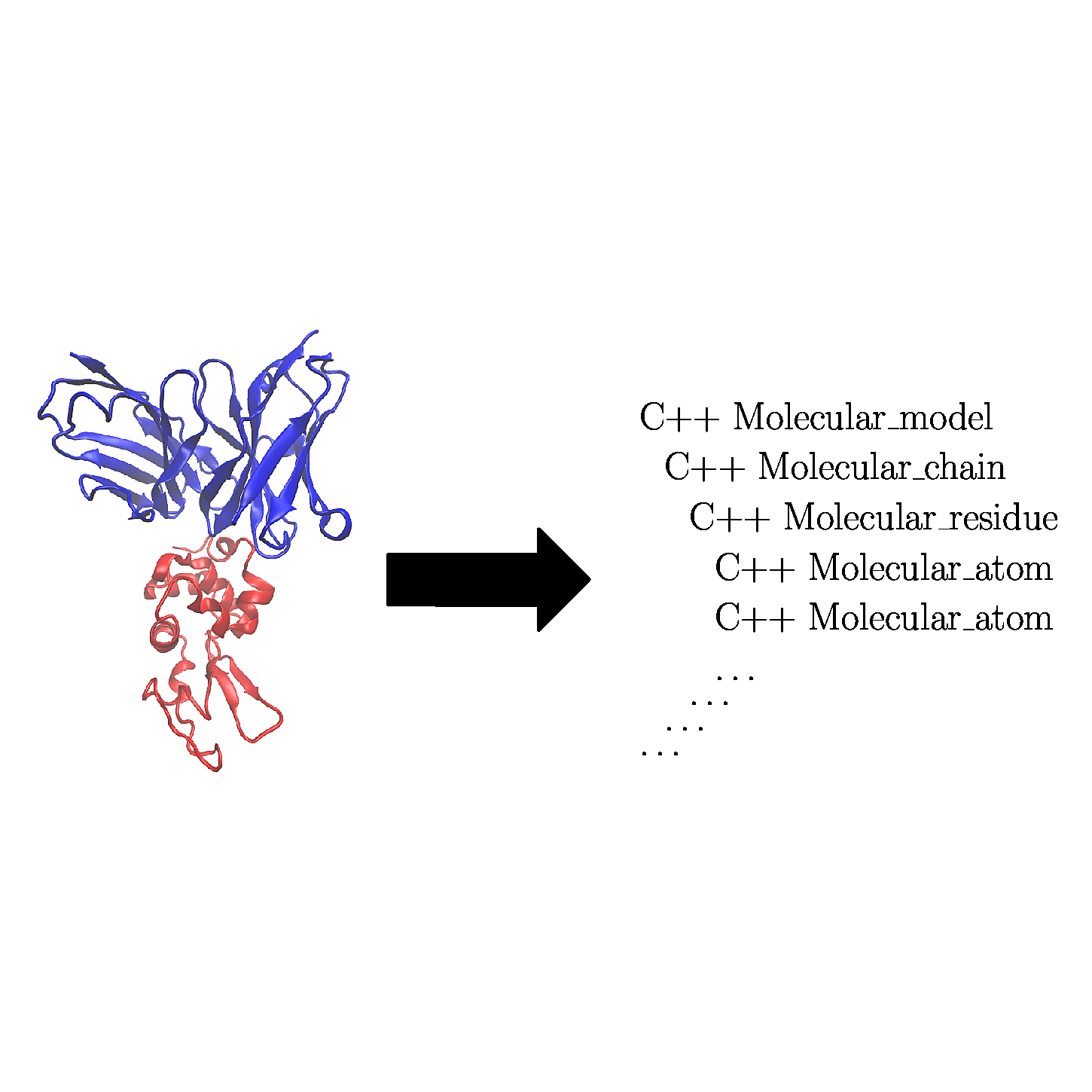 | Concept defining how to load a molecular geometric model from a file. Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus |
| MolecularSystemLabelsTraits | |
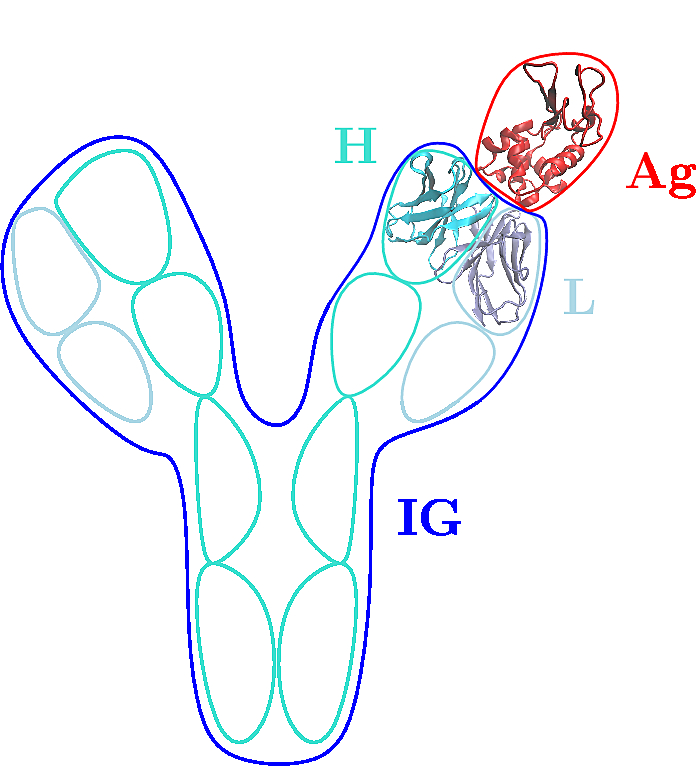 | Concept defining labels to identify particles in a molecular system associated to a molecular structure Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus |
| ParticleAnnotator | |
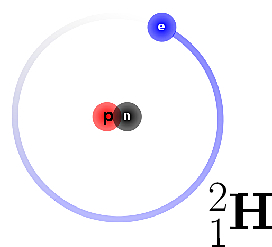 | Concept defining how to decorate particles with supplemental information. Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus |
| ParticleTraits | |
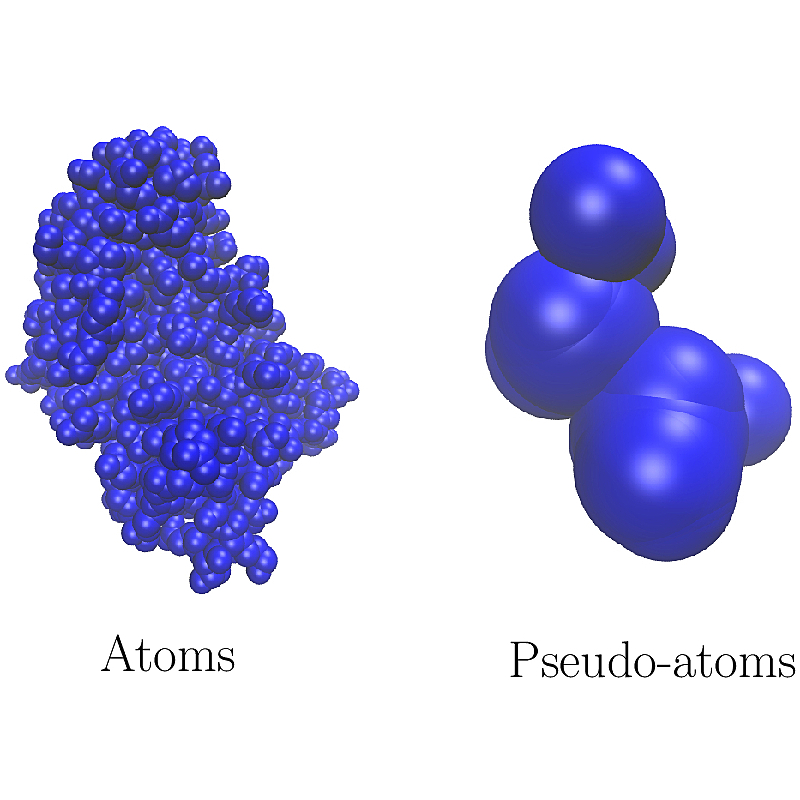 | Concept defining geometric (and possibly biophysical) properties of a particle. Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus |
| UnitSystemTraits | |
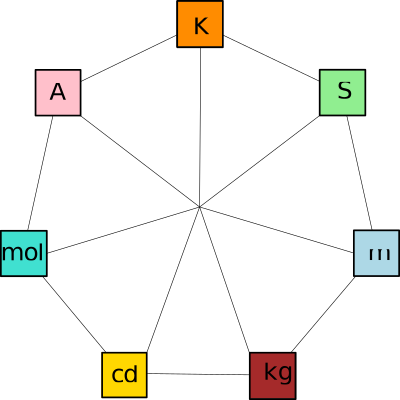 | Concept defining types and constants for physical unit systems Reference Manual – User Manual . Authors: F. Cazals and T. Dreyfus |
Models are instantiations of concepts.
Development in the SBL follows the usual C++ pattern consisting of C++ concepts and C++ models. In a nutshell, C++ concepts define specifications, while C++ models implement such specifications in various contexts. The Models set of packages provide various C++ models to represent molecular systems, but also to perform IO operations.